| Full name: BCAS2 pre-mRNA processing factor | Alias Symbol: DAM1|SPF27|Snt309 | ||
| Type: protein-coding gene | Cytoband: 1p13.2 | ||
| Entrez ID: 10286 | HGNC ID: HGNC:975 | Ensembl Gene: ENSG00000116752 | OMIM ID: 605783 |
| Drug and gene relationship at DGIdb | |||
Expression of BCAS2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BCAS2 | 10286 | 203053_at | -0.5810 | 0.1560 | |
| GSE20347 | BCAS2 | 10286 | 203053_at | 0.1311 | 0.4137 | |
| GSE23400 | BCAS2 | 10286 | 203053_at | 0.0894 | 0.2204 | |
| GSE26886 | BCAS2 | 10286 | 203053_at | 0.1275 | 0.5994 | |
| GSE29001 | BCAS2 | 10286 | 203053_at | 0.1426 | 0.6531 | |
| GSE38129 | BCAS2 | 10286 | 203053_at | 0.1733 | 0.1479 | |
| GSE45670 | BCAS2 | 10286 | 203053_at | 0.0685 | 0.6766 | |
| GSE53622 | BCAS2 | 10286 | 5119 | -0.1217 | 0.0244 | |
| GSE53624 | BCAS2 | 10286 | 5119 | 0.1394 | 0.0136 | |
| GSE63941 | BCAS2 | 10286 | 203053_at | 0.2692 | 0.7121 | |
| GSE77861 | BCAS2 | 10286 | 203053_at | -0.1220 | 0.5657 | |
| GSE97050 | BCAS2 | 10286 | A_23_P34930 | 0.0063 | 0.9805 | |
| SRP007169 | BCAS2 | 10286 | RNAseq | 0.3677 | 0.3663 | |
| SRP008496 | BCAS2 | 10286 | RNAseq | 0.1281 | 0.6667 | |
| SRP064894 | BCAS2 | 10286 | RNAseq | -0.0942 | 0.6431 | |
| SRP133303 | BCAS2 | 10286 | RNAseq | 0.2407 | 0.1552 | |
| SRP159526 | BCAS2 | 10286 | RNAseq | -0.6773 | 0.0002 | |
| SRP193095 | BCAS2 | 10286 | RNAseq | -0.2152 | 0.1005 | |
| SRP219564 | BCAS2 | 10286 | RNAseq | -0.3036 | 0.5700 | |
| TCGA | BCAS2 | 10286 | RNAseq | 0.0860 | 0.1242 |
Upregulated datasets: 0; Downregulated datasets: 0.
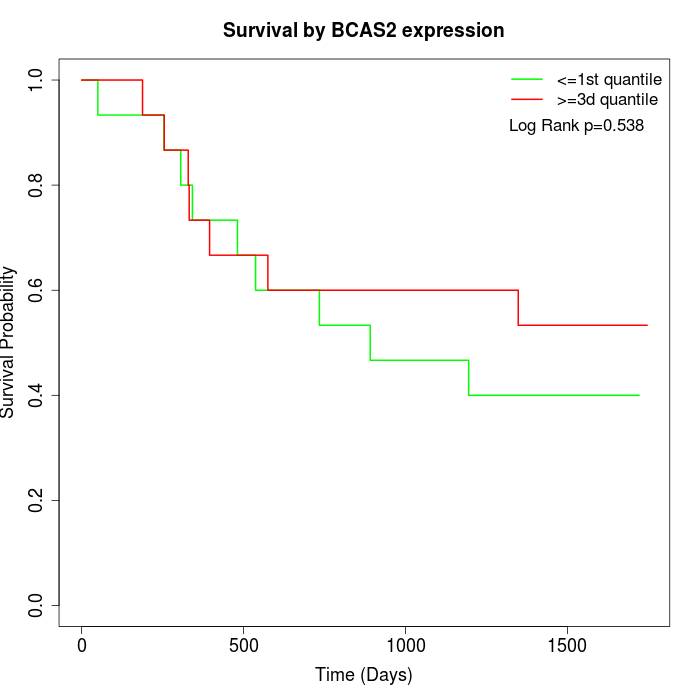
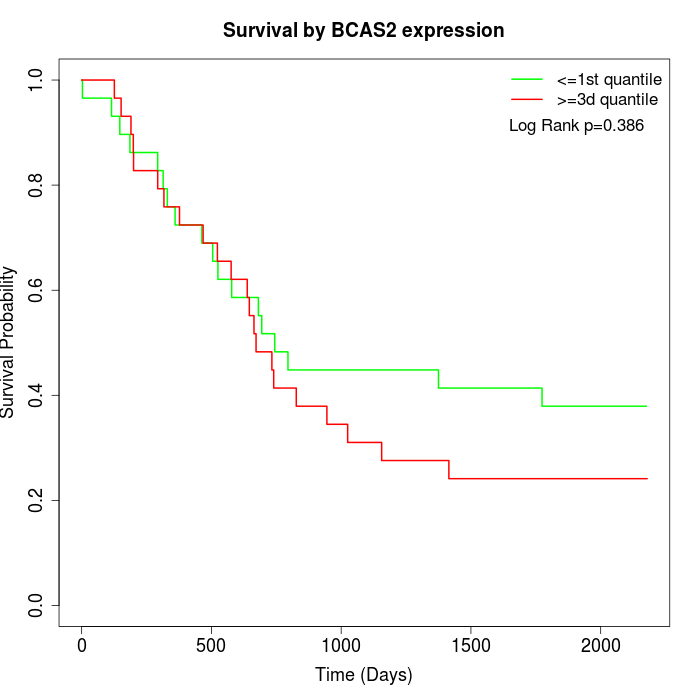
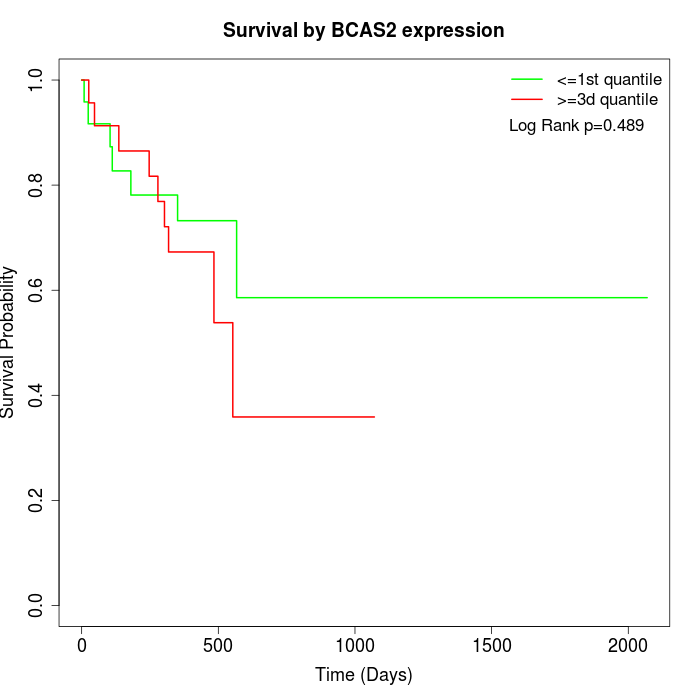
Survival by BCAS2 expression:
Note: Click image to view full size file.
Copy number change of BCAS2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BCAS2 | 10286 | 0 | 9 | 21 | |
| GSE20123 | BCAS2 | 10286 | 0 | 9 | 21 | |
| GSE43470 | BCAS2 | 10286 | 0 | 8 | 35 | |
| GSE46452 | BCAS2 | 10286 | 2 | 1 | 56 | |
| GSE47630 | BCAS2 | 10286 | 9 | 5 | 26 | |
| GSE54993 | BCAS2 | 10286 | 0 | 1 | 69 | |
| GSE54994 | BCAS2 | 10286 | 7 | 3 | 43 | |
| GSE60625 | BCAS2 | 10286 | 0 | 0 | 11 | |
| GSE74703 | BCAS2 | 10286 | 0 | 7 | 29 | |
| GSE74704 | BCAS2 | 10286 | 0 | 5 | 15 | |
| TCGA | BCAS2 | 10286 | 12 | 30 | 54 |
Total number of gains: 30; Total number of losses: 78; Total Number of normals: 380.
Somatic mutations of BCAS2:
Generating mutation plots.
Highly correlated genes for BCAS2:
Showing top 20/203 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BCAS2 | CGRRF1 | 0.800931 | 3 | 0 | 3 |
| BCAS2 | PNPLA8 | 0.782956 | 3 | 0 | 3 |
| BCAS2 | ITPR1 | 0.772836 | 3 | 0 | 3 |
| BCAS2 | ANAPC16 | 0.768631 | 3 | 0 | 3 |
| BCAS2 | PPP3CB | 0.764051 | 4 | 0 | 3 |
| BCAS2 | DYNC1LI2 | 0.758172 | 3 | 0 | 3 |
| BCAS2 | TMEM50A | 0.750972 | 3 | 0 | 3 |
| BCAS2 | N6AMT1 | 0.747713 | 3 | 0 | 3 |
| BCAS2 | UAP1 | 0.746136 | 3 | 0 | 3 |
| BCAS2 | TES | 0.742816 | 3 | 0 | 3 |
| BCAS2 | TOB2 | 0.735158 | 3 | 0 | 3 |
| BCAS2 | SGMS2 | 0.731931 | 3 | 0 | 3 |
| BCAS2 | ZNF266 | 0.731601 | 3 | 0 | 3 |
| BCAS2 | HSPA2 | 0.730963 | 3 | 0 | 3 |
| BCAS2 | LNPEP | 0.729321 | 3 | 0 | 3 |
| BCAS2 | SAMD8 | 0.726307 | 3 | 0 | 3 |
| BCAS2 | PPP2R3C | 0.722871 | 3 | 0 | 3 |
| BCAS2 | TMEM106B | 0.720475 | 4 | 0 | 3 |
| BCAS2 | SNTB1 | 0.71335 | 3 | 0 | 3 |
| BCAS2 | CAMTA1 | 0.711549 | 3 | 0 | 3 |
For details and further investigation, click here