| Full name: heat shock protein family A (Hsp70) member 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 14q23.3 | ||
| Entrez ID: 3306 | HGNC ID: HGNC:5235 | Ensembl Gene: ENSG00000126803 | OMIM ID: 140560 |
| Drug and gene relationship at DGIdb | |||
HSPA2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04213 | Longevity regulating pathway - multiple species | |
| hsa05134 | Legionellosis | |
| hsa05145 | Toxoplasmosis | |
| hsa05164 | Influenza A |
Expression of HSPA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HSPA2 | 3306 | 211538_s_at | -0.8637 | 0.4609 | |
| GSE20347 | HSPA2 | 3306 | 211538_s_at | -0.9313 | 0.0069 | |
| GSE23400 | HSPA2 | 3306 | 211538_s_at | -0.5582 | 0.0125 | |
| GSE26886 | HSPA2 | 3306 | 211538_s_at | -0.3486 | 0.4206 | |
| GSE29001 | HSPA2 | 3306 | 211538_s_at | -0.1935 | 0.7459 | |
| GSE38129 | HSPA2 | 3306 | 211538_s_at | -1.1473 | 0.0001 | |
| GSE45670 | HSPA2 | 3306 | 211538_s_at | -0.8953 | 0.0281 | |
| GSE53622 | HSPA2 | 3306 | 148250 | -0.8801 | 0.0000 | |
| GSE53624 | HSPA2 | 3306 | 148250 | -0.9786 | 0.0000 | |
| GSE63941 | HSPA2 | 3306 | 211538_s_at | 0.0580 | 0.9716 | |
| GSE77861 | HSPA2 | 3306 | 211538_s_at | -0.9888 | 0.2751 | |
| GSE97050 | HSPA2 | 3306 | A_23_P88303 | -0.2718 | 0.4919 | |
| SRP007169 | HSPA2 | 3306 | RNAseq | -1.2383 | 0.0964 | |
| SRP008496 | HSPA2 | 3306 | RNAseq | -1.7485 | 0.0011 | |
| SRP064894 | HSPA2 | 3306 | RNAseq | -0.2798 | 0.3509 | |
| SRP133303 | HSPA2 | 3306 | RNAseq | -0.5522 | 0.1885 | |
| SRP193095 | HSPA2 | 3306 | RNAseq | -0.0389 | 0.9097 | |
| SRP219564 | HSPA2 | 3306 | RNAseq | -0.7794 | 0.2645 | |
| TCGA | HSPA2 | 3306 | RNAseq | -0.0401 | 0.7869 |
Upregulated datasets: 0; Downregulated datasets: 2.
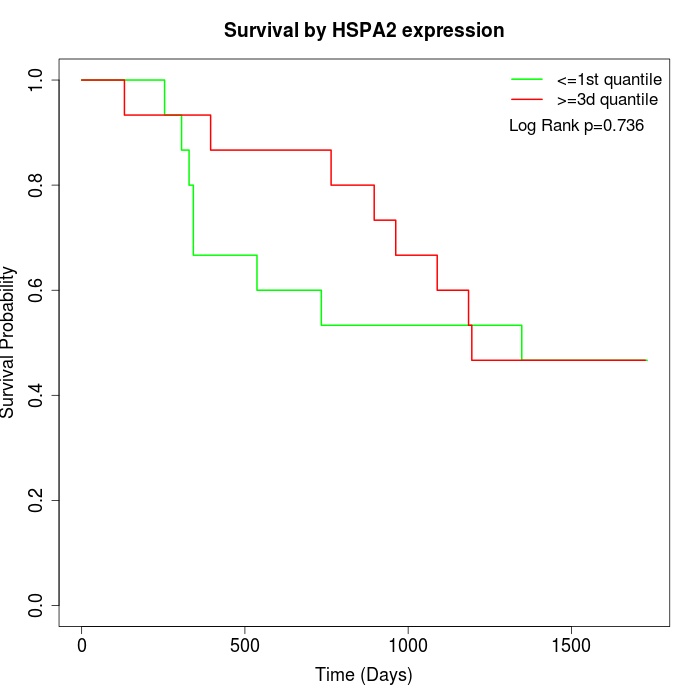
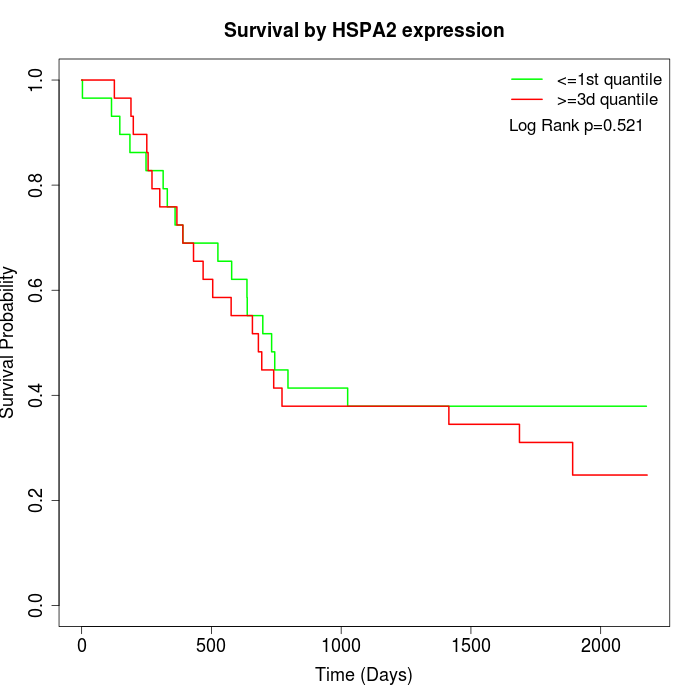
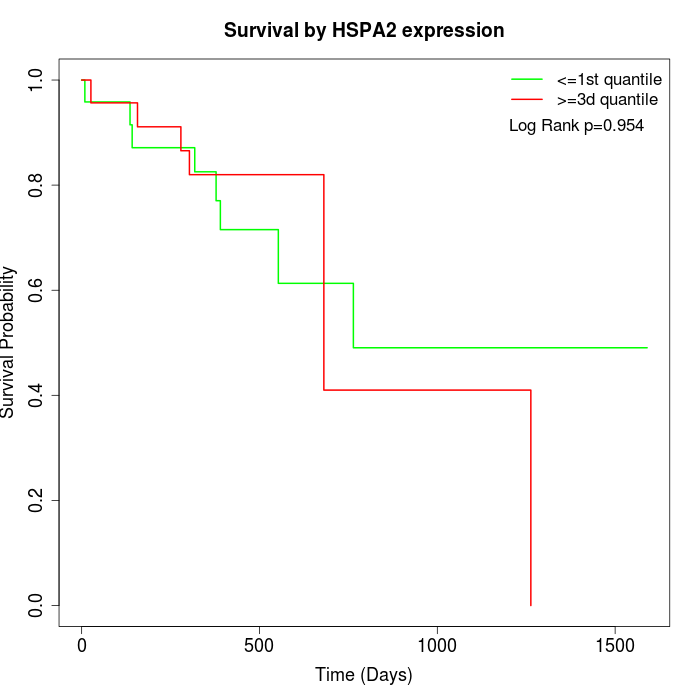
Survival by HSPA2 expression:
Note: Click image to view full size file.
Copy number change of HSPA2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HSPA2 | 3306 | 7 | 3 | 20 | |
| GSE20123 | HSPA2 | 3306 | 7 | 3 | 20 | |
| GSE43470 | HSPA2 | 3306 | 8 | 2 | 33 | |
| GSE46452 | HSPA2 | 3306 | 16 | 3 | 40 | |
| GSE47630 | HSPA2 | 3306 | 11 | 9 | 20 | |
| GSE54993 | HSPA2 | 3306 | 3 | 9 | 58 | |
| GSE54994 | HSPA2 | 3306 | 18 | 5 | 30 | |
| GSE60625 | HSPA2 | 3306 | 0 | 2 | 9 | |
| GSE74703 | HSPA2 | 3306 | 7 | 2 | 27 | |
| GSE74704 | HSPA2 | 3306 | 3 | 2 | 15 | |
| TCGA | HSPA2 | 3306 | 32 | 14 | 50 |
Total number of gains: 112; Total number of losses: 54; Total Number of normals: 322.
Somatic mutations of HSPA2:
Generating mutation plots.
Highly correlated genes for HSPA2:
Showing top 20/227 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HSPA2 | MIER3 | 0.784435 | 3 | 0 | 3 |
| HSPA2 | PITPNM3 | 0.778031 | 3 | 0 | 3 |
| HSPA2 | BBS9 | 0.741941 | 3 | 0 | 3 |
| HSPA2 | BCAS2 | 0.730963 | 3 | 0 | 3 |
| HSPA2 | TRIM23 | 0.719011 | 3 | 0 | 3 |
| HSPA2 | ZFAND6 | 0.711874 | 3 | 0 | 3 |
| HSPA2 | MRPL20 | 0.711693 | 3 | 0 | 3 |
| HSPA2 | SKAP2 | 0.711689 | 4 | 0 | 3 |
| HSPA2 | SSBP3 | 0.711592 | 3 | 0 | 3 |
| HSPA2 | NEK9 | 0.707069 | 4 | 0 | 3 |
| HSPA2 | GLT8D2 | 0.703397 | 4 | 0 | 3 |
| HSPA2 | GNG5 | 0.687274 | 3 | 0 | 3 |
| HSPA2 | RPL41 | 0.685831 | 3 | 0 | 3 |
| HSPA2 | ATL2 | 0.676511 | 3 | 0 | 3 |
| HSPA2 | CRK | 0.667004 | 3 | 0 | 3 |
| HSPA2 | MBLAC2 | 0.664297 | 3 | 0 | 3 |
| HSPA2 | ZFP36L1 | 0.661018 | 4 | 0 | 4 |
| HSPA2 | GSTO2 | 0.659059 | 4 | 0 | 3 |
| HSPA2 | ANKRD13A | 0.657102 | 3 | 0 | 3 |
| HSPA2 | SSU72 | 0.656737 | 3 | 0 | 3 |
For details and further investigation, click here