| Full name: BMI1 proto-oncogene, polycomb ring finger | Alias Symbol: RNF51 | ||
| Type: protein-coding gene | Cytoband: 10p12.2 | ||
| Entrez ID: 648 | HGNC ID: HGNC:1066 | Ensembl Gene: ENSG00000168283 | OMIM ID: 164831 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of BMI1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | BMI1 | 648 | 31084 | 0.1897 | 0.0369 | |
| GSE53624 | BMI1 | 648 | 31084 | 0.2159 | 0.0109 | |
| GSE97050 | BMI1 | 648 | A_24_P303989 | 0.1443 | 0.6914 | |
| SRP007169 | BMI1 | 648 | RNAseq | 0.1588 | 0.8835 | |
| SRP008496 | BMI1 | 648 | RNAseq | 0.1484 | 0.8776 | |
| SRP064894 | BMI1 | 648 | RNAseq | -0.1411 | 0.7686 | |
| SRP133303 | BMI1 | 648 | RNAseq | 0.1379 | 0.3942 | |
| SRP159526 | BMI1 | 648 | RNAseq | -0.1933 | 0.5053 | |
| SRP193095 | BMI1 | 648 | RNAseq | 0.0960 | 0.6013 | |
| SRP219564 | BMI1 | 648 | RNAseq | -0.4219 | 0.4136 | |
| TCGA | BMI1 | 648 | RNAseq | -0.0345 | 0.4917 |
Upregulated datasets: 0; Downregulated datasets: 0.
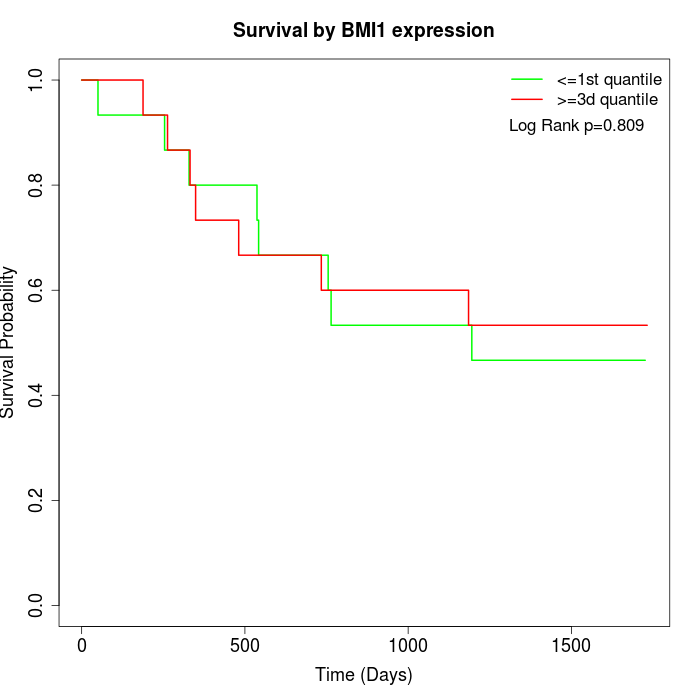
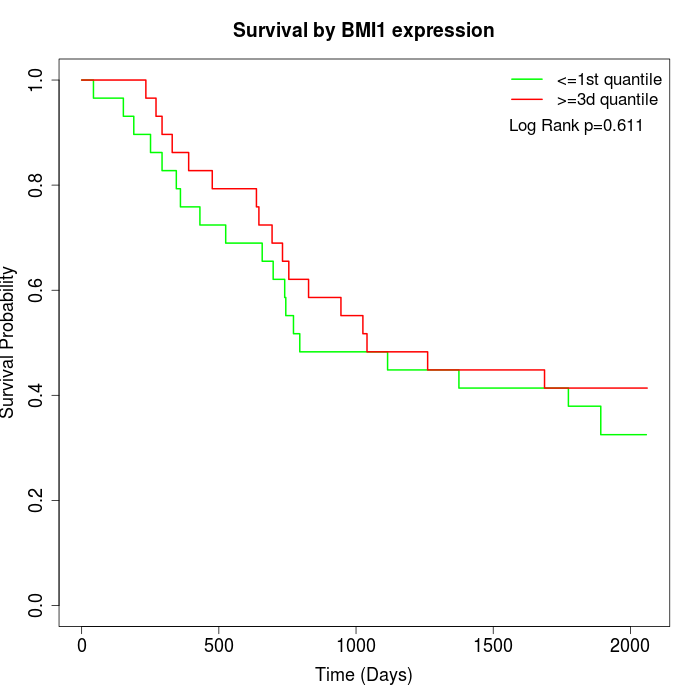
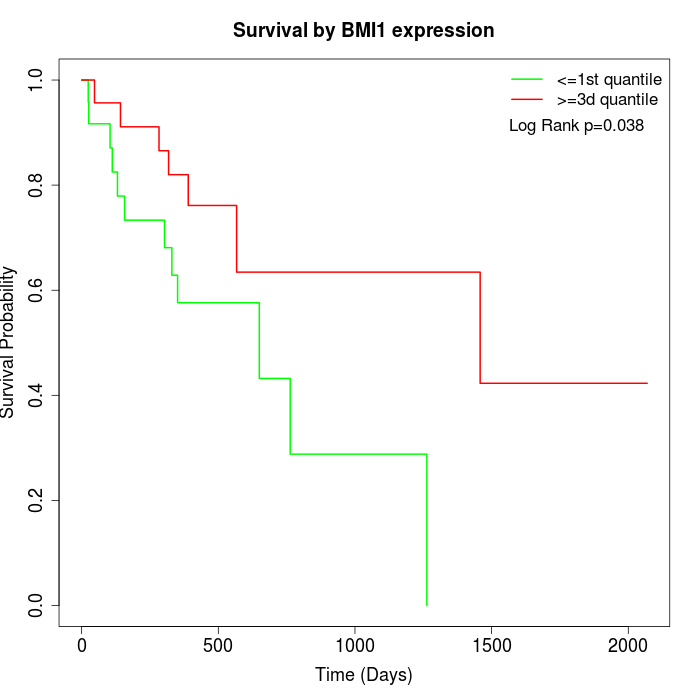
Survival by BMI1 expression:
Note: Click image to view full size file.
Copy number change of BMI1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BMI1 | 648 | 5 | 8 | 17 | |
| GSE20123 | BMI1 | 648 | 5 | 7 | 18 | |
| GSE43470 | BMI1 | 648 | 4 | 4 | 35 | |
| GSE46452 | BMI1 | 648 | 1 | 14 | 44 | |
| GSE47630 | BMI1 | 648 | 5 | 15 | 20 | |
| GSE54993 | BMI1 | 648 | 10 | 0 | 60 | |
| GSE54994 | BMI1 | 648 | 3 | 9 | 41 | |
| GSE60625 | BMI1 | 648 | 0 | 0 | 11 | |
| GSE74703 | BMI1 | 648 | 3 | 2 | 31 | |
| GSE74704 | BMI1 | 648 | 0 | 6 | 14 | |
| TCGA | BMI1 | 648 | 18 | 25 | 53 |
Total number of gains: 54; Total number of losses: 90; Total Number of normals: 344.
Somatic mutations of BMI1:
Generating mutation plots.
Highly correlated genes for BMI1:
Showing top 20/59 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BMI1 | PPP1CC | 0.726173 | 3 | 0 | 3 |
| BMI1 | RCBTB1 | 0.723777 | 3 | 0 | 3 |
| BMI1 | UBR1 | 0.711372 | 3 | 0 | 3 |
| BMI1 | TNS3 | 0.702886 | 3 | 0 | 3 |
| BMI1 | MAP4K3 | 0.702779 | 3 | 0 | 3 |
| BMI1 | PRDX3 | 0.698198 | 3 | 0 | 3 |
| BMI1 | MRAS | 0.697567 | 3 | 0 | 3 |
| BMI1 | CEP120 | 0.695846 | 3 | 0 | 3 |
| BMI1 | HSD11B1 | 0.687657 | 3 | 0 | 3 |
| BMI1 | MRPL33 | 0.687021 | 3 | 0 | 3 |
| BMI1 | RRAGB | 0.683761 | 3 | 0 | 3 |
| BMI1 | SPRED1 | 0.683681 | 3 | 0 | 3 |
| BMI1 | RNASEH2B | 0.682124 | 3 | 0 | 3 |
| BMI1 | SMURF2 | 0.680853 | 3 | 0 | 3 |
| BMI1 | MAT2B | 0.678731 | 3 | 0 | 3 |
| BMI1 | ATF6 | 0.67754 | 3 | 0 | 3 |
| BMI1 | SLC25A38 | 0.67572 | 3 | 0 | 3 |
| BMI1 | ZNF227 | 0.67268 | 3 | 0 | 3 |
| BMI1 | ALG10 | 0.671408 | 3 | 0 | 3 |
| BMI1 | CCDC82 | 0.670077 | 3 | 0 | 3 |
For details and further investigation, click here