| Full name: carbonic anhydrase 12 | Alias Symbol: HsT18816 | ||
| Type: protein-coding gene | Cytoband: 15q22.2 | ||
| Entrez ID: 771 | HGNC ID: HGNC:1371 | Ensembl Gene: ENSG00000074410 | OMIM ID: 603263 |
| Drug and gene relationship at DGIdb | |||
Expression of CA12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CA12 | 771 | 215867_x_at | -0.1732 | 0.8266 | |
| GSE20347 | CA12 | 771 | 203963_at | -0.3260 | 0.3424 | |
| GSE23400 | CA12 | 771 | 203963_at | -0.1753 | 0.3411 | |
| GSE26886 | CA12 | 771 | 203963_at | -0.9401 | 0.0050 | |
| GSE29001 | CA12 | 771 | 214164_x_at | -0.3523 | 0.4172 | |
| GSE38129 | CA12 | 771 | 203963_at | 0.1431 | 0.8374 | |
| GSE45670 | CA12 | 771 | 215867_x_at | 0.1024 | 0.5966 | |
| GSE53622 | CA12 | 771 | 13169 | -0.3436 | 0.2284 | |
| GSE53624 | CA12 | 771 | 13169 | -0.3960 | 0.0076 | |
| GSE63941 | CA12 | 771 | 215867_x_at | -2.4152 | 0.0120 | |
| GSE77861 | CA12 | 771 | 203963_at | 0.2961 | 0.3839 | |
| GSE97050 | CA12 | 771 | A_24_P330518 | 0.0062 | 0.9916 | |
| SRP007169 | CA12 | 771 | RNAseq | -0.3501 | 0.5576 | |
| SRP008496 | CA12 | 771 | RNAseq | -0.5480 | 0.1923 | |
| SRP064894 | CA12 | 771 | RNAseq | 0.0713 | 0.8349 | |
| SRP133303 | CA12 | 771 | RNAseq | -0.1146 | 0.7765 | |
| SRP159526 | CA12 | 771 | RNAseq | -0.6587 | 0.3760 | |
| SRP193095 | CA12 | 771 | RNAseq | 0.2403 | 0.4989 | |
| SRP219564 | CA12 | 771 | RNAseq | 0.3281 | 0.7445 | |
| TCGA | CA12 | 771 | RNAseq | 0.3861 | 0.0046 |
Upregulated datasets: 0; Downregulated datasets: 1.
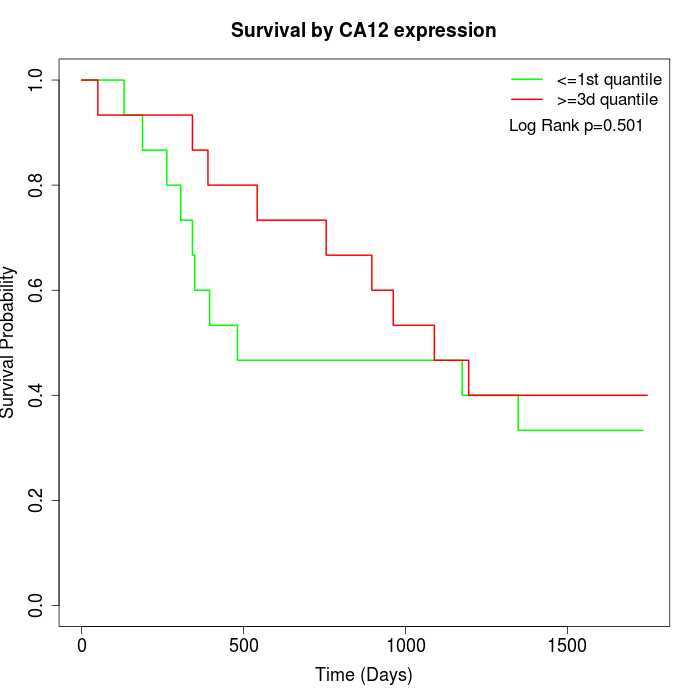
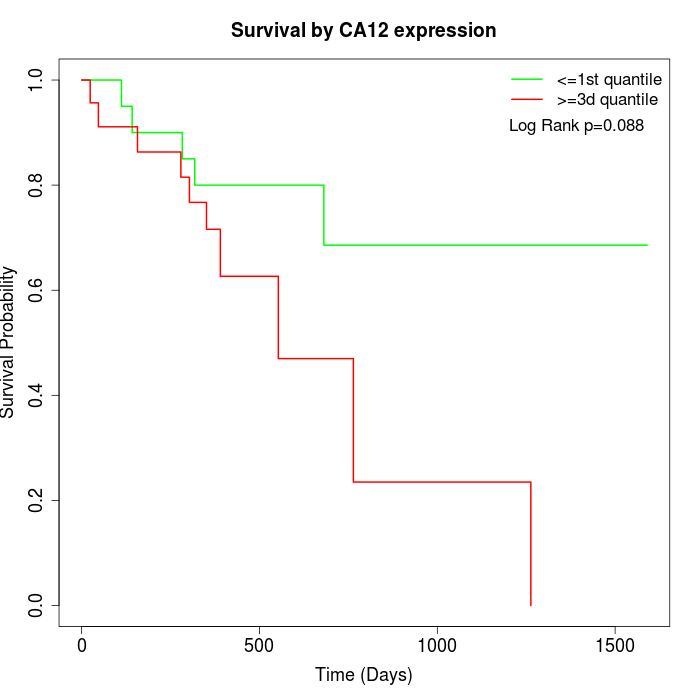
Survival by CA12 expression:
Note: Click image to view full size file.
Copy number change of CA12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CA12 | 771 | 8 | 2 | 20 | |
| GSE20123 | CA12 | 771 | 8 | 2 | 20 | |
| GSE43470 | CA12 | 771 | 4 | 6 | 33 | |
| GSE46452 | CA12 | 771 | 3 | 7 | 49 | |
| GSE47630 | CA12 | 771 | 8 | 10 | 22 | |
| GSE54993 | CA12 | 771 | 5 | 6 | 59 | |
| GSE54994 | CA12 | 771 | 6 | 8 | 39 | |
| GSE60625 | CA12 | 771 | 4 | 0 | 7 | |
| GSE74703 | CA12 | 771 | 4 | 3 | 29 | |
| GSE74704 | CA12 | 771 | 4 | 2 | 14 | |
| TCGA | CA12 | 771 | 10 | 17 | 69 |
Total number of gains: 64; Total number of losses: 63; Total Number of normals: 361.
Somatic mutations of CA12:
Generating mutation plots.
Highly correlated genes for CA12:
Showing top 20/255 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CA12 | ZNF770 | 0.712944 | 4 | 0 | 4 |
| CA12 | MAP4K5 | 0.685527 | 3 | 0 | 3 |
| CA12 | NOTCH3 | 0.684825 | 4 | 0 | 4 |
| CA12 | DIP2B | 0.673874 | 4 | 0 | 4 |
| CA12 | GOPC | 0.668026 | 3 | 0 | 3 |
| CA12 | KCNK6 | 0.666874 | 7 | 0 | 6 |
| CA12 | SEMA4B | 0.666647 | 5 | 0 | 3 |
| CA12 | BICD2 | 0.660278 | 10 | 0 | 9 |
| CA12 | ZBED6 | 0.659104 | 3 | 0 | 3 |
| CA12 | PLEKHA2 | 0.65877 | 3 | 0 | 3 |
| CA12 | TBC1D2 | 0.656427 | 10 | 0 | 7 |
| CA12 | SPTSSB | 0.655448 | 3 | 0 | 3 |
| CA12 | MCTP1 | 0.655176 | 3 | 0 | 3 |
| CA12 | PGLYRP3 | 0.649928 | 4 | 0 | 3 |
| CA12 | METRNL | 0.648503 | 5 | 0 | 5 |
| CA12 | EXOSC6 | 0.648149 | 3 | 0 | 3 |
| CA12 | PPP2R2C | 0.647677 | 6 | 0 | 4 |
| CA12 | CHMP2B | 0.646624 | 3 | 0 | 3 |
| CA12 | IPMK | 0.644369 | 5 | 0 | 4 |
| CA12 | CDK5R1 | 0.642715 | 8 | 0 | 6 |
For details and further investigation, click here