| Full name: caveolin 2 | Alias Symbol: CAV | ||
| Type: protein-coding gene | Cytoband: 7q31.2 | ||
| Entrez ID: 858 | HGNC ID: HGNC:1528 | Ensembl Gene: ENSG00000105971 | OMIM ID: 601048 |
| Drug and gene relationship at DGIdb | |||
CAV2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04510 | Focal adhesion | |
| hsa05205 | Proteoglycans in cancer |
Expression of CAV2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CAV2 | 858 | 203324_s_at | 0.0693 | 0.9470 | |
| GSE20347 | CAV2 | 858 | 203324_s_at | 0.6088 | 0.1339 | |
| GSE23400 | CAV2 | 858 | 203323_at | 0.3411 | 0.0444 | |
| GSE26886 | CAV2 | 858 | 203323_at | 0.7635 | 0.0347 | |
| GSE29001 | CAV2 | 858 | 203324_s_at | 1.0764 | 0.0303 | |
| GSE38129 | CAV2 | 858 | 203324_s_at | 0.5139 | 0.0769 | |
| GSE45670 | CAV2 | 858 | 203324_s_at | 0.4891 | 0.0958 | |
| GSE53622 | CAV2 | 858 | 53687 | 0.7894 | 0.0000 | |
| GSE53624 | CAV2 | 858 | 103660 | 1.1030 | 0.0000 | |
| GSE63941 | CAV2 | 858 | 203324_s_at | -0.4813 | 0.5790 | |
| GSE77861 | CAV2 | 858 | 203324_s_at | 0.8588 | 0.0219 | |
| GSE97050 | CAV2 | 858 | A_23_P123071 | 0.4501 | 0.1903 | |
| SRP007169 | CAV2 | 858 | RNAseq | 1.0400 | 0.0350 | |
| SRP008496 | CAV2 | 858 | RNAseq | 1.3507 | 0.0000 | |
| SRP064894 | CAV2 | 858 | RNAseq | 0.9361 | 0.0000 | |
| SRP133303 | CAV2 | 858 | RNAseq | 0.9709 | 0.0000 | |
| SRP159526 | CAV2 | 858 | RNAseq | 0.0484 | 0.9159 | |
| SRP193095 | CAV2 | 858 | RNAseq | 0.9230 | 0.0001 | |
| SRP219564 | CAV2 | 858 | RNAseq | 0.4529 | 0.2957 | |
| TCGA | CAV2 | 858 | RNAseq | 0.2428 | 0.0038 |
Upregulated datasets: 4; Downregulated datasets: 0.
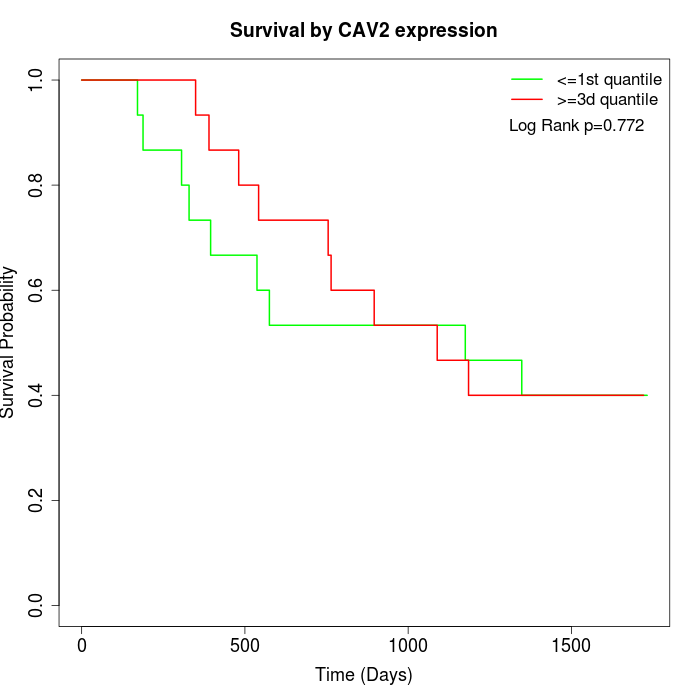
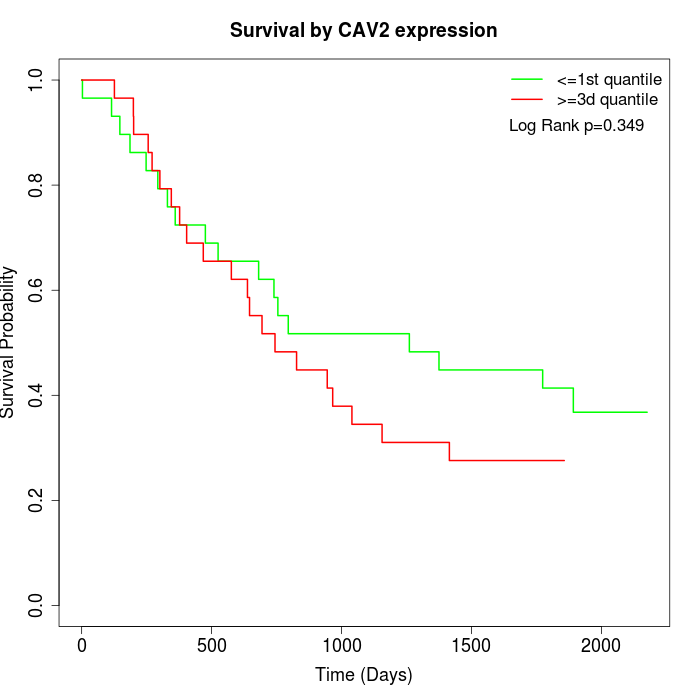
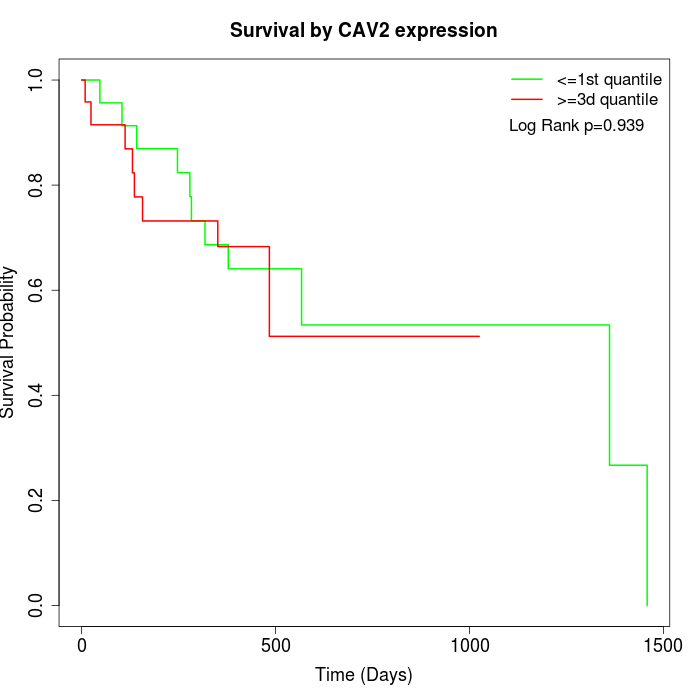
Survival by CAV2 expression:
Note: Click image to view full size file.
Copy number change of CAV2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CAV2 | 858 | 10 | 2 | 18 | |
| GSE20123 | CAV2 | 858 | 11 | 2 | 17 | |
| GSE43470 | CAV2 | 858 | 3 | 2 | 38 | |
| GSE46452 | CAV2 | 858 | 8 | 1 | 50 | |
| GSE47630 | CAV2 | 858 | 7 | 4 | 29 | |
| GSE54993 | CAV2 | 858 | 2 | 7 | 61 | |
| GSE54994 | CAV2 | 858 | 11 | 6 | 36 | |
| GSE60625 | CAV2 | 858 | 0 | 0 | 11 | |
| GSE74703 | CAV2 | 858 | 3 | 2 | 31 | |
| GSE74704 | CAV2 | 858 | 7 | 2 | 11 | |
| TCGA | CAV2 | 858 | 33 | 17 | 46 |
Total number of gains: 95; Total number of losses: 45; Total Number of normals: 348.
Somatic mutations of CAV2:
Generating mutation plots.
Highly correlated genes for CAV2:
Showing top 20/1188 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CAV2 | CAV1 | 0.785067 | 13 | 0 | 13 |
| CAV2 | RNMT | 0.75133 | 3 | 0 | 3 |
| CAV2 | CASC4 | 0.74581 | 3 | 0 | 3 |
| CAV2 | CHMP7 | 0.728782 | 3 | 0 | 3 |
| CAV2 | DGKH | 0.726251 | 6 | 0 | 6 |
| CAV2 | PPP1R21 | 0.721796 | 4 | 0 | 3 |
| CAV2 | GNG10 | 0.72174 | 3 | 0 | 3 |
| CAV2 | RLIM | 0.717423 | 3 | 0 | 3 |
| CAV2 | SNRNP200 | 0.714801 | 3 | 0 | 3 |
| CAV2 | SPON2 | 0.711007 | 3 | 0 | 3 |
| CAV2 | TRIM35 | 0.709419 | 3 | 0 | 3 |
| CAV2 | WDR81 | 0.706375 | 3 | 0 | 3 |
| CAV2 | CCP110 | 0.705537 | 4 | 0 | 3 |
| CAV2 | FBLIM1 | 0.703717 | 6 | 0 | 5 |
| CAV2 | ZC3HC1 | 0.702477 | 3 | 0 | 3 |
| CAV2 | SGTB | 0.7006 | 5 | 0 | 5 |
| CAV2 | C6orf141 | 0.698169 | 5 | 0 | 4 |
| CAV2 | FANCL | 0.697674 | 3 | 0 | 3 |
| CAV2 | TMEM138 | 0.692961 | 5 | 0 | 4 |
| CAV2 | EVL | 0.69116 | 3 | 0 | 3 |
For details and further investigation, click here