| Full name: cyclin E1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 19q12 | ||
| Entrez ID: 898 | HGNC ID: HGNC:1589 | Ensembl Gene: ENSG00000105173 | OMIM ID: 123837 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CCNE1 involved pathways:
Expression of CCNE1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCNE1 | 898 | 213523_at | 1.5640 | 0.0186 | |
| GSE20347 | CCNE1 | 898 | 213523_at | 0.7258 | 0.0002 | |
| GSE23400 | CCNE1 | 898 | 213523_at | 0.3971 | 0.0001 | |
| GSE26886 | CCNE1 | 898 | 213523_at | 0.5445 | 0.0109 | |
| GSE29001 | CCNE1 | 898 | 213523_at | 1.0519 | 0.0022 | |
| GSE38129 | CCNE1 | 898 | 213523_at | 0.9035 | 0.0000 | |
| GSE45670 | CCNE1 | 898 | 213523_at | 1.0885 | 0.0002 | |
| GSE53622 | CCNE1 | 898 | 57509 | 0.6861 | 0.0000 | |
| GSE53624 | CCNE1 | 898 | 57509 | 1.0744 | 0.0000 | |
| GSE63941 | CCNE1 | 898 | 213523_at | -0.1794 | 0.8320 | |
| GSE77861 | CCNE1 | 898 | 213523_at | 0.5361 | 0.1287 | |
| GSE97050 | CCNE1 | 898 | A_23_P209200 | 0.5359 | 0.2416 | |
| SRP007169 | CCNE1 | 898 | RNAseq | 0.0462 | 0.9408 | |
| SRP008496 | CCNE1 | 898 | RNAseq | 0.1649 | 0.6709 | |
| SRP064894 | CCNE1 | 898 | RNAseq | 1.0269 | 0.0010 | |
| SRP133303 | CCNE1 | 898 | RNAseq | 1.0235 | 0.0004 | |
| SRP159526 | CCNE1 | 898 | RNAseq | 1.4267 | 0.0010 | |
| SRP193095 | CCNE1 | 898 | RNAseq | 0.6228 | 0.0001 | |
| SRP219564 | CCNE1 | 898 | RNAseq | 0.5612 | 0.3241 | |
| TCGA | CCNE1 | 898 | RNAseq | 0.8392 | 0.0000 |
Upregulated datasets: 7; Downregulated datasets: 0.
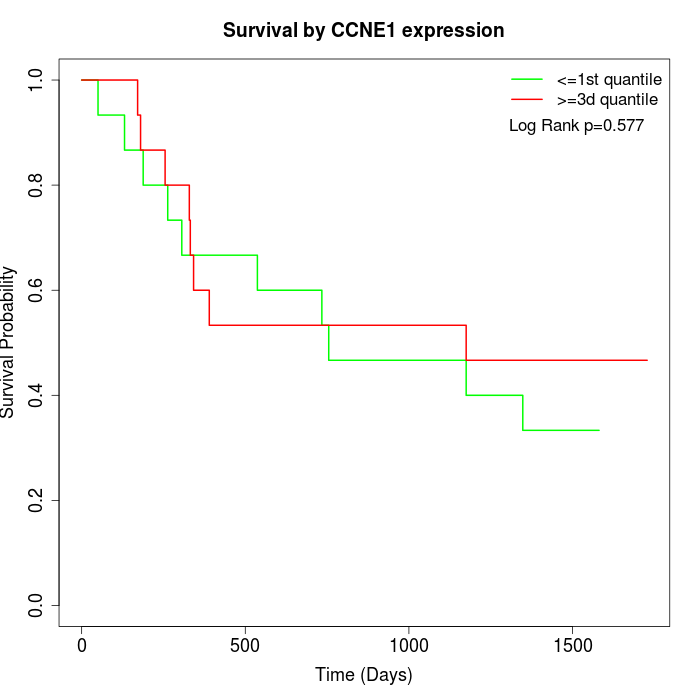
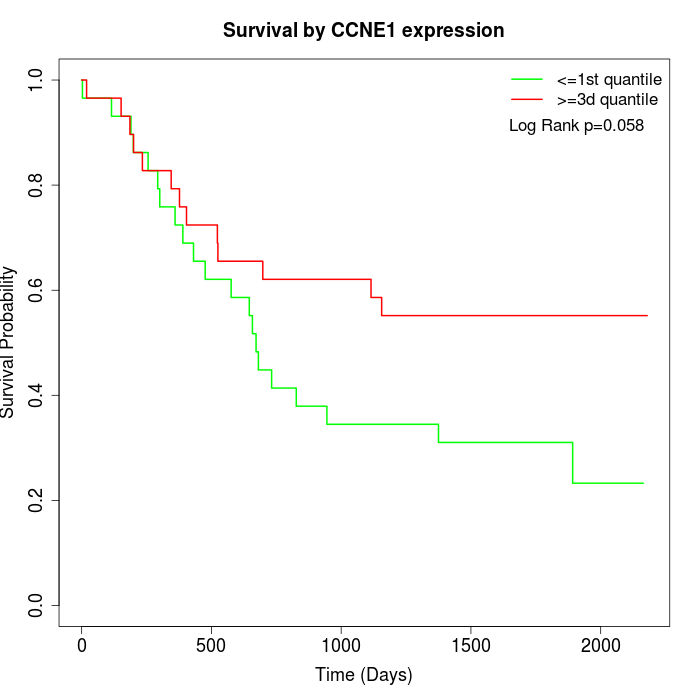
Survival by CCNE1 expression:
Note: Click image to view full size file.
Copy number change of CCNE1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCNE1 | 898 | 6 | 7 | 17 | |
| GSE20123 | CCNE1 | 898 | 6 | 6 | 18 | |
| GSE43470 | CCNE1 | 898 | 6 | 6 | 31 | |
| GSE46452 | CCNE1 | 898 | 47 | 1 | 11 | |
| GSE47630 | CCNE1 | 898 | 8 | 6 | 26 | |
| GSE54993 | CCNE1 | 898 | 17 | 3 | 50 | |
| GSE54994 | CCNE1 | 898 | 7 | 11 | 35 | |
| GSE60625 | CCNE1 | 898 | 9 | 0 | 2 | |
| GSE74703 | CCNE1 | 898 | 6 | 4 | 26 | |
| GSE74704 | CCNE1 | 898 | 5 | 4 | 11 | |
| TCGA | CCNE1 | 898 | 18 | 12 | 66 |
Total number of gains: 135; Total number of losses: 60; Total Number of normals: 293.
Somatic mutations of CCNE1:
Generating mutation plots.
Highly correlated genes for CCNE1:
Showing top 20/1437 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCNE1 | CDCA2 | 0.788169 | 3 | 0 | 3 |
| CCNE1 | DHX33 | 0.757204 | 4 | 0 | 4 |
| CCNE1 | GORAB | 0.743547 | 4 | 0 | 3 |
| CCNE1 | UBQLN4 | 0.740848 | 4 | 0 | 4 |
| CCNE1 | DOCK6 | 0.736826 | 4 | 0 | 4 |
| CCNE1 | WIPI1 | 0.736807 | 3 | 0 | 3 |
| CCNE1 | UBE2T | 0.727502 | 5 | 0 | 5 |
| CCNE1 | DSCC1 | 0.722523 | 6 | 0 | 5 |
| CCNE1 | STX16 | 0.719468 | 3 | 0 | 3 |
| CCNE1 | LRRC58 | 0.718864 | 4 | 0 | 3 |
| CCNE1 | FABP4 | 0.714741 | 3 | 0 | 3 |
| CCNE1 | ZFYVE27 | 0.710121 | 3 | 0 | 3 |
| CCNE1 | ZNF830 | 0.703687 | 3 | 0 | 3 |
| CCNE1 | EXOC8 | 0.700518 | 3 | 0 | 3 |
| CCNE1 | MRPL14 | 0.699833 | 5 | 0 | 4 |
| CCNE1 | DHX37 | 0.69824 | 6 | 0 | 6 |
| CCNE1 | F2RL1 | 0.69716 | 3 | 0 | 3 |
| CCNE1 | ZAP70 | 0.696679 | 3 | 0 | 3 |
| CCNE1 | MAGOHB | 0.693816 | 6 | 0 | 5 |
| CCNE1 | NACC1 | 0.692607 | 6 | 0 | 5 |
For details and further investigation, click here