| Full name: chaperonin containing TCP1 subunit 2 | Alias Symbol: Cctb | ||
| Type: protein-coding gene | Cytoband: 12q15 | ||
| Entrez ID: 10576 | HGNC ID: HGNC:1615 | Ensembl Gene: ENSG00000166226 | OMIM ID: 605139 |
| Drug and gene relationship at DGIdb | |||
Expression of CCT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCT2 | 10576 | 201947_s_at | 0.5361 | 0.0965 | |
| GSE20347 | CCT2 | 10576 | 201947_s_at | 0.7924 | 0.0000 | |
| GSE23400 | CCT2 | 10576 | 201947_s_at | 0.8959 | 0.0000 | |
| GSE26886 | CCT2 | 10576 | 201947_s_at | 0.3565 | 0.0027 | |
| GSE29001 | CCT2 | 10576 | 201947_s_at | 0.6866 | 0.1872 | |
| GSE38129 | CCT2 | 10576 | 201947_s_at | 0.9100 | 0.0000 | |
| GSE45670 | CCT2 | 10576 | 201947_s_at | 0.4837 | 0.0038 | |
| GSE53622 | CCT2 | 10576 | 71192 | 0.7316 | 0.0000 | |
| GSE53624 | CCT2 | 10576 | 71192 | 0.9963 | 0.0000 | |
| GSE63941 | CCT2 | 10576 | 201947_s_at | 0.5865 | 0.2203 | |
| GSE77861 | CCT2 | 10576 | 201947_s_at | 0.6076 | 0.0163 | |
| GSE97050 | CCT2 | 10576 | A_23_P105392 | 0.5298 | 0.1133 | |
| SRP007169 | CCT2 | 10576 | RNAseq | 0.1045 | 0.7477 | |
| SRP008496 | CCT2 | 10576 | RNAseq | 0.1730 | 0.2998 | |
| SRP064894 | CCT2 | 10576 | RNAseq | 0.5080 | 0.0018 | |
| SRP133303 | CCT2 | 10576 | RNAseq | 1.3898 | 0.0001 | |
| SRP159526 | CCT2 | 10576 | RNAseq | 0.8074 | 0.0483 | |
| SRP193095 | CCT2 | 10576 | RNAseq | 0.3546 | 0.0216 | |
| TCGA | CCT2 | 10576 | RNAseq | 0.2373 | 0.0001 |
Upregulated datasets: 1; Downregulated datasets: 0.
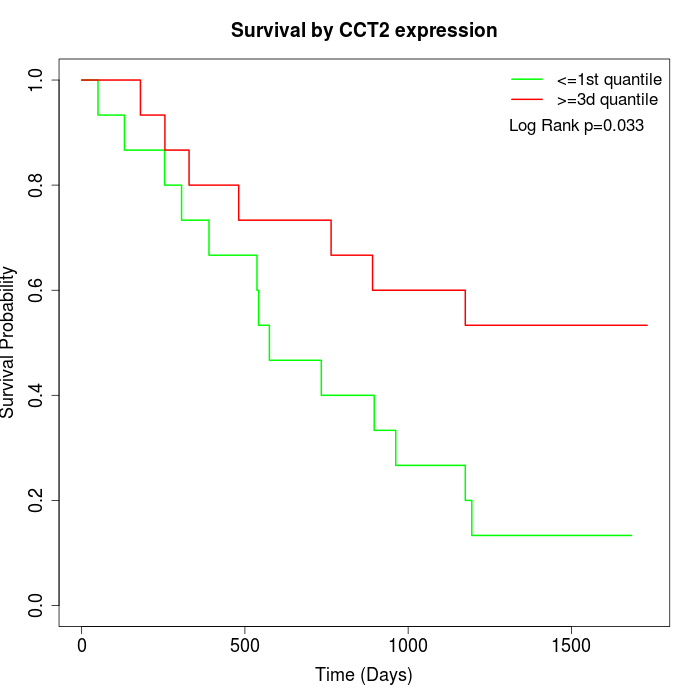
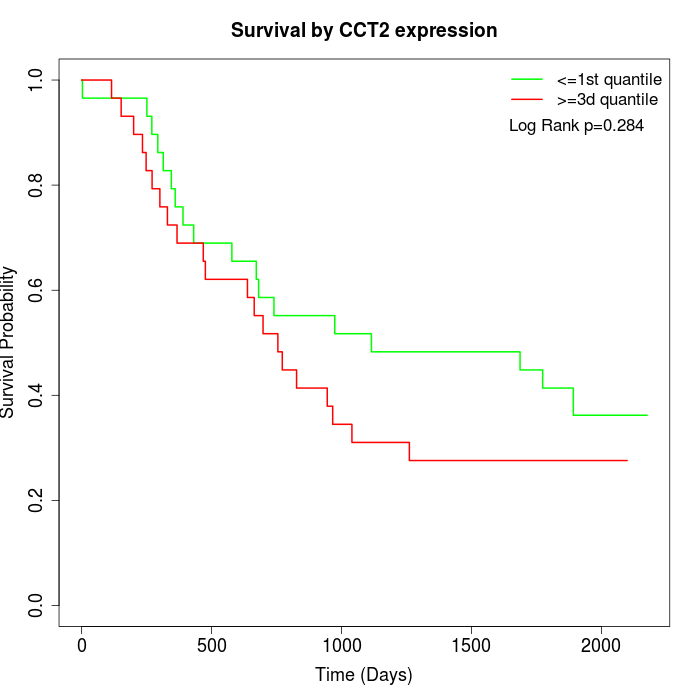
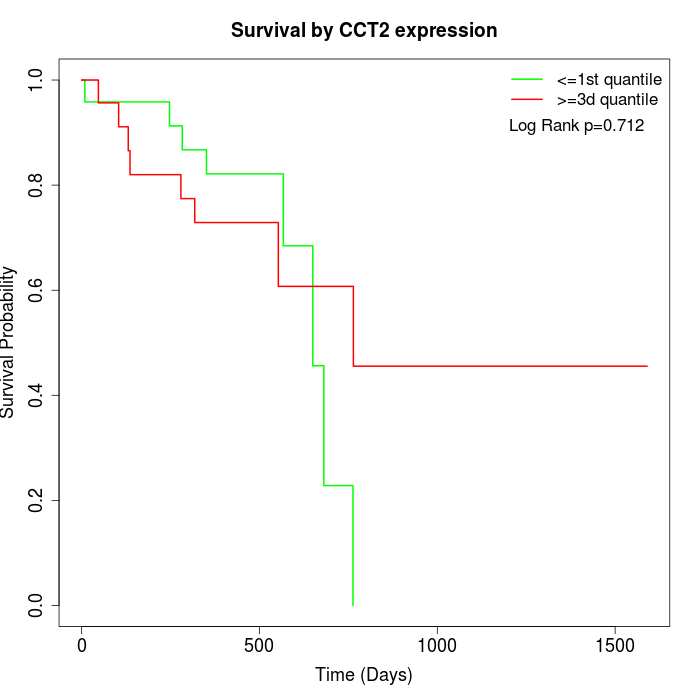
Survival by CCT2 expression:
Note: Click image to view full size file.
Copy number change of CCT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCT2 | 10576 | 6 | 1 | 23 | |
| GSE20123 | CCT2 | 10576 | 6 | 0 | 24 | |
| GSE43470 | CCT2 | 10576 | 4 | 0 | 39 | |
| GSE46452 | CCT2 | 10576 | 11 | 1 | 47 | |
| GSE47630 | CCT2 | 10576 | 10 | 1 | 29 | |
| GSE54993 | CCT2 | 10576 | 0 | 6 | 64 | |
| GSE54994 | CCT2 | 10576 | 8 | 1 | 44 | |
| GSE60625 | CCT2 | 10576 | 0 | 0 | 11 | |
| GSE74703 | CCT2 | 10576 | 4 | 0 | 32 | |
| GSE74704 | CCT2 | 10576 | 5 | 0 | 15 | |
| TCGA | CCT2 | 10576 | 23 | 8 | 65 |
Total number of gains: 77; Total number of losses: 18; Total Number of normals: 393.
Somatic mutations of CCT2:
Generating mutation plots.
Highly correlated genes for CCT2:
Showing top 20/1984 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCT2 | INTS10 | 0.80061 | 3 | 0 | 3 |
| CCT2 | NUP107 | 0.783638 | 13 | 0 | 13 |
| CCT2 | FARS2 | 0.767844 | 3 | 0 | 3 |
| CCT2 | ELP4 | 0.756231 | 3 | 0 | 3 |
| CCT2 | TTL | 0.751096 | 7 | 0 | 7 |
| CCT2 | PYCR2 | 0.750784 | 3 | 0 | 3 |
| CCT2 | EBNA1BP2 | 0.749703 | 4 | 0 | 4 |
| CCT2 | MAPKAPK5 | 0.74448 | 11 | 0 | 11 |
| CCT2 | MMS22L | 0.743915 | 3 | 0 | 3 |
| CCT2 | FADS1 | 0.738933 | 3 | 0 | 3 |
| CCT2 | TRAF7 | 0.737226 | 4 | 0 | 4 |
| CCT2 | MCM6 | 0.733703 | 11 | 0 | 10 |
| CCT2 | RUVBL1 | 0.732499 | 13 | 0 | 13 |
| CCT2 | ILF2 | 0.730462 | 12 | 0 | 11 |
| CCT2 | EFTUD2 | 0.729948 | 7 | 0 | 7 |
| CCT2 | CCT4 | 0.726016 | 12 | 0 | 10 |
| CCT2 | SPON2 | 0.724861 | 3 | 0 | 3 |
| CCT2 | NOP56 | 0.724297 | 3 | 0 | 3 |
| CCT2 | DHRS13 | 0.723243 | 3 | 0 | 3 |
| CCT2 | PTCD1 | 0.72231 | 3 | 0 | 3 |
For details and further investigation, click here