| Full name: complement factor B | Alias Symbol: H2-Bf | ||
| Type: protein-coding gene | Cytoband: 6p21.33 | ||
| Entrez ID: 629 | HGNC ID: HGNC:1037 | Ensembl Gene: ENSG00000243649 | OMIM ID: 138470 |
| Drug and gene relationship at DGIdb | |||
CFB involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04610 | Complement and coagulation cascades | |
| hsa05150 | Staphylococcus aureus infection |
Expression of CFB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CFB | 629 | 202357_s_at | 0.4489 | 0.6782 | |
| GSE20347 | CFB | 629 | 202357_s_at | -0.1141 | 0.7977 | |
| GSE23400 | CFB | 629 | 202357_s_at | 0.1937 | 0.1624 | |
| GSE26886 | CFB | 629 | 202357_s_at | -0.9784 | 0.0696 | |
| GSE29001 | CFB | 629 | 202357_s_at | 0.5277 | 0.3152 | |
| GSE38129 | CFB | 629 | 202357_s_at | -0.0633 | 0.8562 | |
| GSE45670 | CFB | 629 | 202357_s_at | 1.1938 | 0.0050 | |
| GSE63941 | CFB | 629 | 202357_s_at | -1.1861 | 0.5281 | |
| GSE77861 | CFB | 629 | 202357_s_at | 0.1083 | 0.8172 | |
| GSE97050 | CFB | 629 | A_23_P156687 | 1.0362 | 0.2237 | |
| SRP007169 | CFB | 629 | RNAseq | 0.9908 | 0.0776 | |
| SRP008496 | CFB | 629 | RNAseq | 0.2721 | 0.4660 | |
| SRP064894 | CFB | 629 | RNAseq | 1.1774 | 0.0026 | |
| SRP133303 | CFB | 629 | RNAseq | 0.3023 | 0.2830 | |
| SRP159526 | CFB | 629 | RNAseq | 0.6322 | 0.2505 | |
| SRP193095 | CFB | 629 | RNAseq | 0.2822 | 0.3800 | |
| SRP219564 | CFB | 629 | RNAseq | 1.8961 | 0.0016 | |
| TCGA | CFB | 629 | RNAseq | -0.1750 | 0.2229 |
Upregulated datasets: 3; Downregulated datasets: 0.
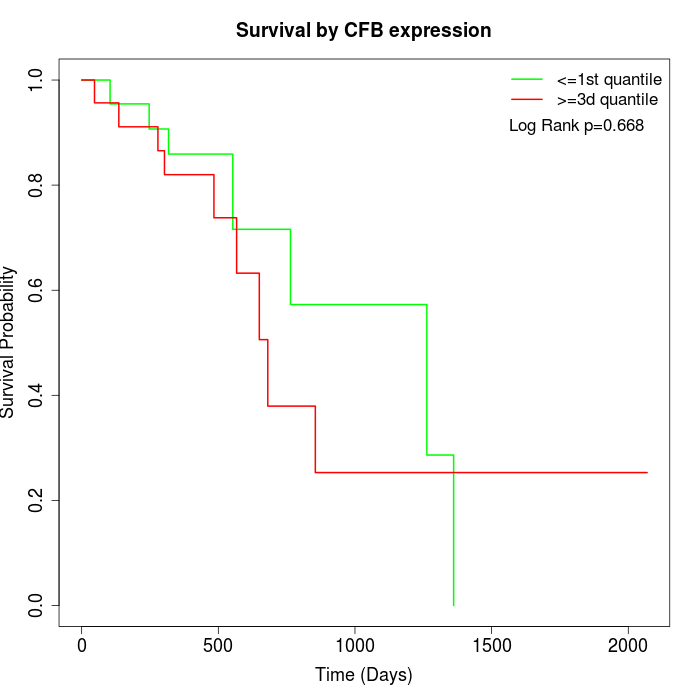
Survival by CFB expression:
Note: Click image to view full size file.
Copy number change of CFB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CFB | 629 | 5 | 1 | 24 | |
| GSE20123 | CFB | 629 | 5 | 1 | 24 | |
| GSE43470 | CFB | 629 | 5 | 1 | 37 | |
| GSE46452 | CFB | 629 | 1 | 10 | 48 | |
| GSE47630 | CFB | 629 | 7 | 4 | 29 | |
| GSE54993 | CFB | 629 | 2 | 1 | 67 | |
| GSE54994 | CFB | 629 | 11 | 4 | 38 | |
| GSE60625 | CFB | 629 | 0 | 1 | 10 | |
| GSE74703 | CFB | 629 | 5 | 0 | 31 | |
| GSE74704 | CFB | 629 | 2 | 0 | 18 | |
| TCGA | CFB | 629 | 16 | 16 | 64 |
Total number of gains: 59; Total number of losses: 39; Total Number of normals: 390.
Somatic mutations of CFB:
Generating mutation plots.
Highly correlated genes for CFB:
Showing top 20/190 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CFB | SLA2 | 0.785836 | 3 | 0 | 3 |
| CFB | DTNB | 0.707907 | 3 | 0 | 3 |
| CFB | MCOLN2 | 0.685239 | 3 | 0 | 3 |
| CFB | MDK | 0.664389 | 4 | 0 | 3 |
| CFB | GBP3 | 0.658424 | 4 | 0 | 4 |
| CFB | LTF | 0.652334 | 4 | 0 | 4 |
| CFB | TRAT1 | 0.64289 | 5 | 0 | 3 |
| CFB | IL2RB | 0.638546 | 6 | 0 | 4 |
| CFB | STAT4 | 0.637631 | 6 | 0 | 5 |
| CFB | TIGIT | 0.635907 | 3 | 0 | 3 |
| CFB | IL10RA | 0.632603 | 7 | 0 | 6 |
| CFB | GZMB | 0.623319 | 9 | 0 | 7 |
| CFB | EVI2A | 0.62127 | 7 | 0 | 6 |
| CFB | IDO1 | 0.618888 | 8 | 0 | 6 |
| CFB | APOL3 | 0.616692 | 9 | 0 | 8 |
| CFB | CYTIP | 0.61184 | 6 | 0 | 5 |
| CFB | MYO1G | 0.611123 | 5 | 0 | 3 |
| CFB | CCL5 | 0.608215 | 6 | 0 | 5 |
| CFB | CD84 | 0.606839 | 4 | 0 | 4 |
| CFB | SIGLEC10 | 0.603568 | 4 | 0 | 3 |
For details and further investigation, click here