| Full name: complement factor H related 3 | Alias Symbol: FHR-3|HLF4|FHR3|DOWN16 | ||
| Type: protein-coding gene | Cytoband: 1q31.3 | ||
| Entrez ID: 10878 | HGNC ID: HGNC:16980 | Ensembl Gene: ENSG00000116785 | OMIM ID: 605336 |
| Drug and gene relationship at DGIdb | |||
Expression of CFHR3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CFHR3 | 10878 | 1554459_s_at | 0.1163 | 0.7077 | |
| GSE26886 | CFHR3 | 10878 | 1554459_s_at | 0.5612 | 0.0034 | |
| GSE45670 | CFHR3 | 10878 | 1554459_s_at | 0.2946 | 0.0214 | |
| GSE53622 | CFHR3 | 10878 | 24084 | -0.7807 | 0.0000 | |
| GSE53624 | CFHR3 | 10878 | 24084 | -0.9040 | 0.0000 | |
| GSE63941 | CFHR3 | 10878 | 1554459_s_at | 0.3843 | 0.1448 | |
| GSE77861 | CFHR3 | 10878 | 1554459_s_at | -0.1284 | 0.4314 | |
| GSE97050 | CFHR3 | 10878 | A_23_P103256 | 0.2622 | 0.4784 | |
| TCGA | CFHR3 | 10878 | RNAseq | 2.1554 | 0.0001 |
Upregulated datasets: 1; Downregulated datasets: 0.
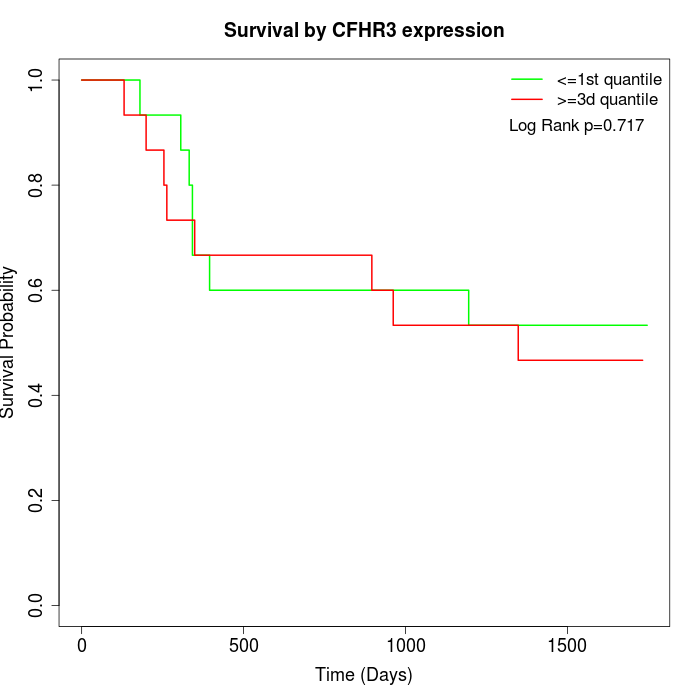
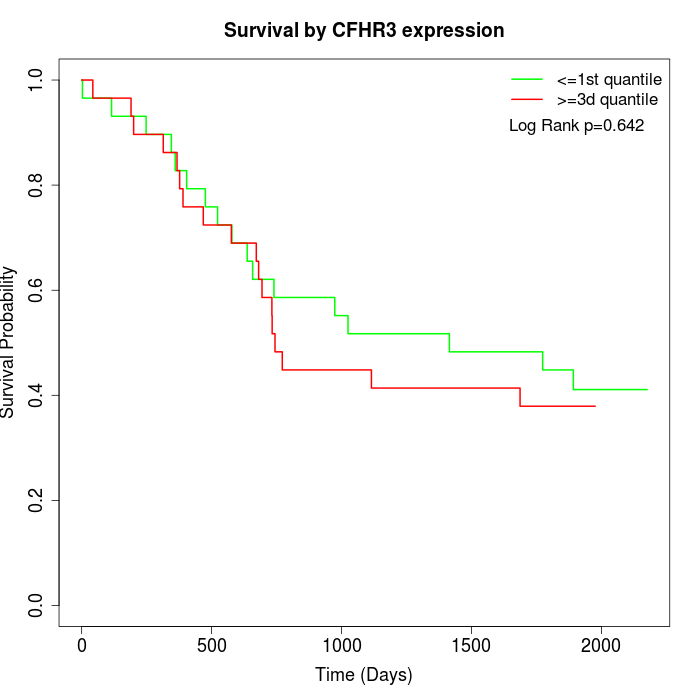
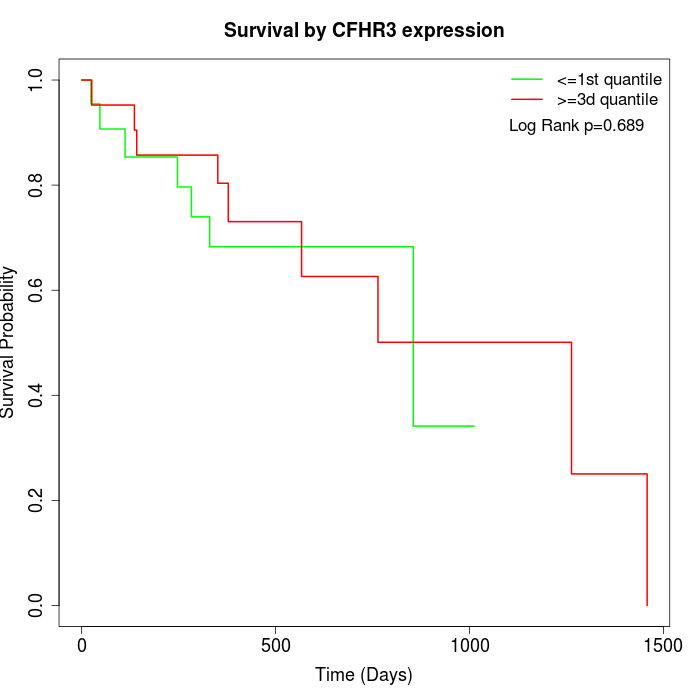
Survival by CFHR3 expression:
Note: Click image to view full size file.
Copy number change of CFHR3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CFHR3 | 10878 | 6 | 1 | 23 | |
| GSE20123 | CFHR3 | 10878 | 6 | 1 | 23 | |
| GSE43470 | CFHR3 | 10878 | 5 | 1 | 37 | |
| GSE46452 | CFHR3 | 10878 | 3 | 1 | 55 | |
| GSE47630 | CFHR3 | 10878 | 14 | 0 | 26 | |
| GSE54993 | CFHR3 | 10878 | 0 | 6 | 64 | |
| GSE54994 | CFHR3 | 10878 | 16 | 0 | 37 | |
| GSE60625 | CFHR3 | 10878 | 0 | 0 | 11 | |
| GSE74703 | CFHR3 | 10878 | 5 | 1 | 30 | |
| GSE74704 | CFHR3 | 10878 | 2 | 1 | 17 | |
| TCGA | CFHR3 | 10878 | 41 | 6 | 49 |
Total number of gains: 98; Total number of losses: 18; Total Number of normals: 372.
Somatic mutations of CFHR3:
Generating mutation plots.
Highly correlated genes for CFHR3:
Showing top 20/87 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CFHR3 | GIMAP5 | 0.802233 | 3 | 0 | 3 |
| CFHR3 | ALKBH6 | 0.766073 | 3 | 0 | 3 |
| CFHR3 | CFDP1 | 0.745028 | 3 | 0 | 3 |
| CFHR3 | CCT8 | 0.744084 | 3 | 0 | 3 |
| CFHR3 | SEC24D | 0.723324 | 3 | 0 | 3 |
| CFHR3 | STX18 | 0.722355 | 3 | 0 | 3 |
| CFHR3 | MRFAP1 | 0.721488 | 3 | 0 | 3 |
| CFHR3 | ARHGAP15 | 0.713484 | 3 | 0 | 3 |
| CFHR3 | GALM | 0.710825 | 3 | 0 | 3 |
| CFHR3 | TBC1D8 | 0.701596 | 4 | 0 | 4 |
| CFHR3 | UBA2 | 0.70059 | 3 | 0 | 3 |
| CFHR3 | EXOSC8 | 0.691124 | 3 | 0 | 3 |
| CFHR3 | EPN1 | 0.687399 | 3 | 0 | 3 |
| CFHR3 | SNRNP35 | 0.685002 | 3 | 0 | 3 |
| CFHR3 | MRPS28 | 0.684635 | 3 | 0 | 3 |
| CFHR3 | TNFSF12 | 0.682037 | 3 | 0 | 3 |
| CFHR3 | CFI | 0.678296 | 4 | 0 | 3 |
| CFHR3 | OSBPL7 | 0.67717 | 3 | 0 | 3 |
| CFHR3 | IFI16 | 0.674985 | 3 | 0 | 3 |
| CFHR3 | TMEM92 | 0.673332 | 3 | 0 | 3 |
For details and further investigation, click here