| Full name: cingulin | Alias Symbol: KIAA1319 | ||
| Type: protein-coding gene | Cytoband: 1q21.3 | ||
| Entrez ID: 57530 | HGNC ID: HGNC:17429 | Ensembl Gene: ENSG00000143375 | OMIM ID: 609473 |
| Drug and gene relationship at DGIdb | |||
CGN involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04530 | Tight junction |
Expression of CGN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CGN | 57530 | 223233_s_at | -0.3415 | 0.6438 | |
| GSE26886 | CGN | 57530 | 223233_s_at | -1.1354 | 0.0024 | |
| GSE45670 | CGN | 57530 | 223233_s_at | -0.1795 | 0.4919 | |
| GSE53622 | CGN | 57530 | 126088 | -1.2605 | 0.0000 | |
| GSE53624 | CGN | 57530 | 126088 | -1.9920 | 0.0000 | |
| GSE63941 | CGN | 57530 | 223233_s_at | 1.5752 | 0.0783 | |
| GSE77861 | CGN | 57530 | 223233_s_at | -0.0912 | 0.7860 | |
| GSE97050 | CGN | 57530 | A_33_P3329974 | -0.1082 | 0.7677 | |
| SRP007169 | CGN | 57530 | RNAseq | -0.1421 | 0.7976 | |
| SRP064894 | CGN | 57530 | RNAseq | -2.2607 | 0.0000 | |
| SRP133303 | CGN | 57530 | RNAseq | -1.3389 | 0.0022 | |
| SRP159526 | CGN | 57530 | RNAseq | -0.9116 | 0.0992 | |
| SRP193095 | CGN | 57530 | RNAseq | -0.7180 | 0.0710 | |
| SRP219564 | CGN | 57530 | RNAseq | -1.5762 | 0.0484 | |
| TCGA | CGN | 57530 | RNAseq | -0.6293 | 0.0006 |
Upregulated datasets: 0; Downregulated datasets: 6.
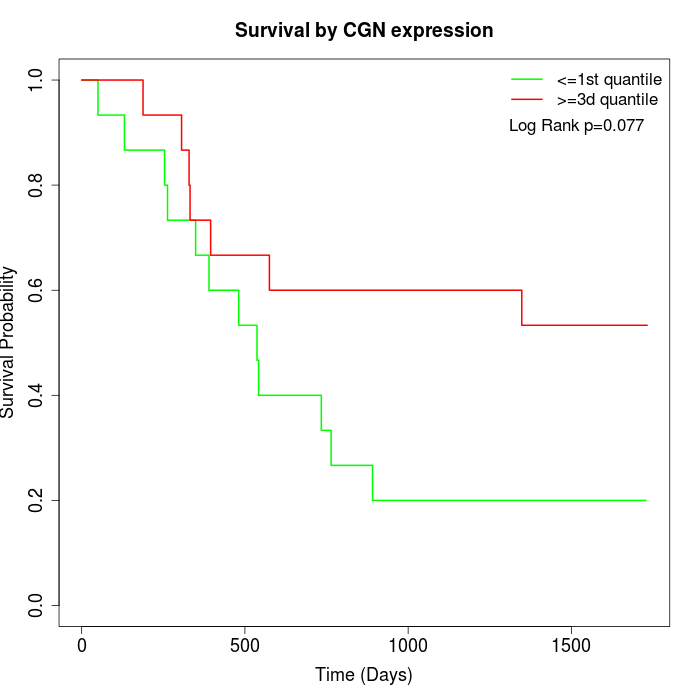
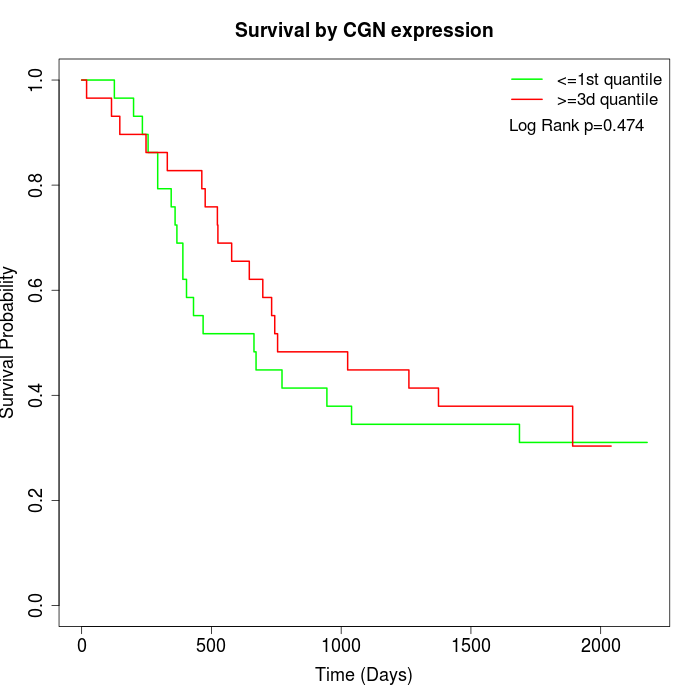
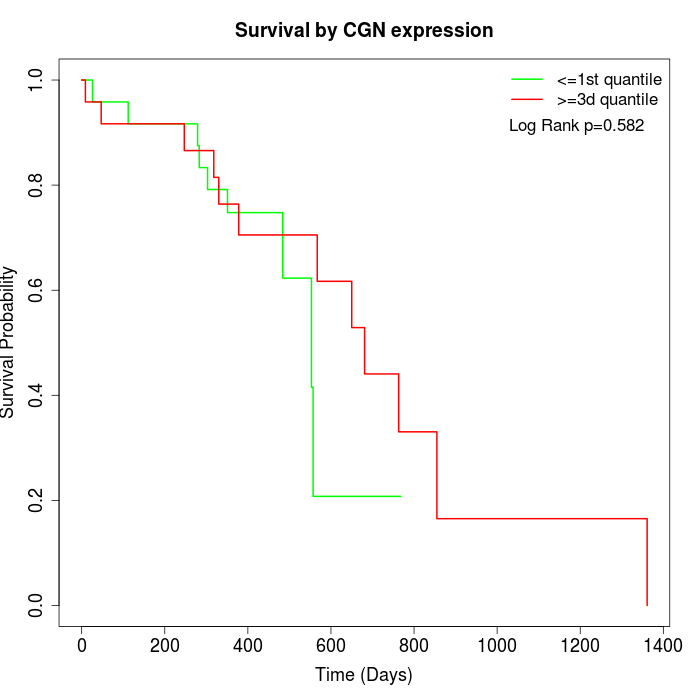
Survival by CGN expression:
Note: Click image to view full size file.
Copy number change of CGN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CGN | 57530 | 15 | 0 | 15 | |
| GSE20123 | CGN | 57530 | 15 | 0 | 15 | |
| GSE43470 | CGN | 57530 | 6 | 1 | 36 | |
| GSE46452 | CGN | 57530 | 2 | 1 | 56 | |
| GSE47630 | CGN | 57530 | 14 | 0 | 26 | |
| GSE54993 | CGN | 57530 | 0 | 4 | 66 | |
| GSE54994 | CGN | 57530 | 15 | 0 | 38 | |
| GSE60625 | CGN | 57530 | 0 | 0 | 11 | |
| GSE74703 | CGN | 57530 | 6 | 1 | 29 | |
| GSE74704 | CGN | 57530 | 7 | 0 | 13 | |
| TCGA | CGN | 57530 | 38 | 2 | 56 |
Total number of gains: 118; Total number of losses: 9; Total Number of normals: 361.
Somatic mutations of CGN:
Generating mutation plots.
Highly correlated genes for CGN:
Showing top 20/856 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CGN | NIPAL1 | 0.74766 | 3 | 0 | 3 |
| CGN | TSPAN6 | 0.723669 | 7 | 0 | 7 |
| CGN | USP6NL | 0.723225 | 6 | 0 | 6 |
| CGN | LTA4H | 0.714097 | 3 | 0 | 3 |
| CGN | ARHGAP27 | 0.713253 | 8 | 0 | 7 |
| CGN | SLC10A5 | 0.711287 | 3 | 0 | 3 |
| CGN | RER1 | 0.708742 | 4 | 0 | 4 |
| CGN | RAB25 | 0.708001 | 8 | 0 | 7 |
| CGN | TST | 0.705148 | 7 | 0 | 6 |
| CGN | TM7SF2 | 0.70336 | 8 | 0 | 7 |
| CGN | ULK3 | 0.697837 | 6 | 0 | 6 |
| CGN | DAPP1 | 0.688053 | 5 | 0 | 5 |
| CGN | RBM47 | 0.68736 | 7 | 0 | 7 |
| CGN | GRHL1 | 0.68735 | 8 | 0 | 8 |
| CGN | CHMP4C | 0.686705 | 7 | 0 | 6 |
| CGN | UPK3B | 0.685924 | 3 | 0 | 3 |
| CGN | SH3BGRL3 | 0.685655 | 3 | 0 | 3 |
| CGN | NUAK2 | 0.68354 | 3 | 0 | 3 |
| CGN | GCNT3 | 0.683464 | 7 | 0 | 7 |
| CGN | ANXA9 | 0.683289 | 7 | 0 | 7 |
For details and further investigation, click here