| Full name: epithelial cell adhesion molecule | Alias Symbol: Ly74|TROP1|GA733-2|EGP34|EGP40|EGP-2|KSA|CD326|Ep-CAM|HEA125|KS1/4|MK-1|MH99|MOC31|323/A3|17-1A|TACST-1|CO-17A|ESA | ||
| Type: protein-coding gene | Cytoband: 2p21 | ||
| Entrez ID: 4072 | HGNC ID: HGNC:11529 | Ensembl Gene: ENSG00000119888 | OMIM ID: 185535 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of EPCAM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EPCAM | 4072 | 201839_s_at | 1.3372 | 0.0864 | |
| GSE20347 | EPCAM | 4072 | 201839_s_at | 2.5773 | 0.0000 | |
| GSE23400 | EPCAM | 4072 | 201839_s_at | 2.0627 | 0.0000 | |
| GSE26886 | EPCAM | 4072 | 201839_s_at | 1.9560 | 0.0001 | |
| GSE29001 | EPCAM | 4072 | 201839_s_at | 1.5384 | 0.0002 | |
| GSE38129 | EPCAM | 4072 | 201839_s_at | 2.7312 | 0.0000 | |
| GSE45670 | EPCAM | 4072 | 201839_s_at | 1.2757 | 0.0449 | |
| GSE53622 | EPCAM | 4072 | 21083 | 2.0033 | 0.0000 | |
| GSE53624 | EPCAM | 4072 | 21083 | 1.7775 | 0.0000 | |
| GSE63941 | EPCAM | 4072 | 201839_s_at | 8.1276 | 0.0000 | |
| GSE77861 | EPCAM | 4072 | 201839_s_at | 2.4776 | 0.0003 | |
| GSE97050 | EPCAM | 4072 | A_23_P91081 | 2.3369 | 0.0631 | |
| SRP007169 | EPCAM | 4072 | RNAseq | 3.8024 | 0.0000 | |
| SRP008496 | EPCAM | 4072 | RNAseq | 4.3183 | 0.0000 | |
| SRP064894 | EPCAM | 4072 | RNAseq | 0.9868 | 0.0365 | |
| SRP133303 | EPCAM | 4072 | RNAseq | 1.8256 | 0.0001 | |
| SRP159526 | EPCAM | 4072 | RNAseq | 3.7540 | 0.0000 | |
| SRP193095 | EPCAM | 4072 | RNAseq | -0.1207 | 0.8081 | |
| SRP219564 | EPCAM | 4072 | RNAseq | 2.2107 | 0.0268 | |
| TCGA | EPCAM | 4072 | RNAseq | 0.1502 | 0.3461 |
Upregulated datasets: 15; Downregulated datasets: 0.
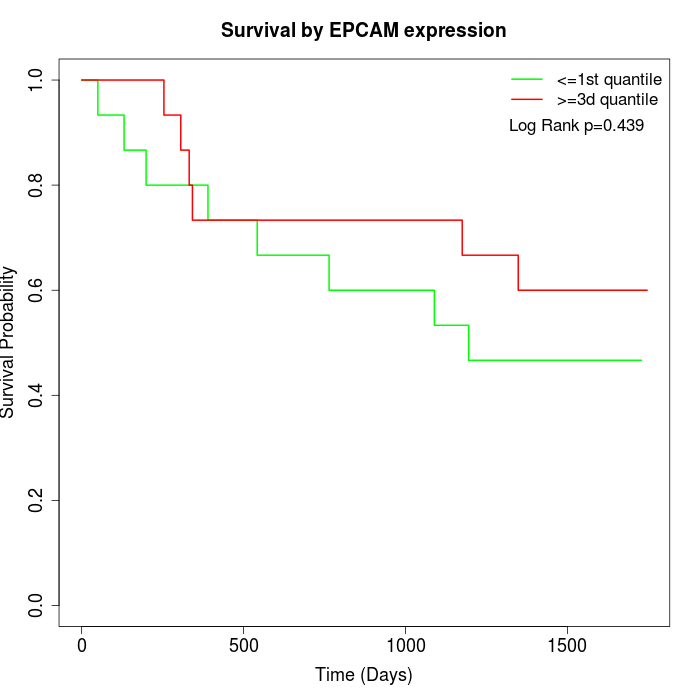
Survival by EPCAM expression:
Note: Click image to view full size file.
Copy number change of EPCAM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EPCAM | 4072 | 10 | 1 | 19 | |
| GSE20123 | EPCAM | 4072 | 8 | 1 | 21 | |
| GSE43470 | EPCAM | 4072 | 5 | 0 | 38 | |
| GSE46452 | EPCAM | 4072 | 1 | 4 | 54 | |
| GSE47630 | EPCAM | 4072 | 8 | 0 | 32 | |
| GSE54993 | EPCAM | 4072 | 0 | 5 | 65 | |
| GSE54994 | EPCAM | 4072 | 12 | 0 | 41 | |
| GSE60625 | EPCAM | 4072 | 0 | 3 | 8 | |
| GSE74703 | EPCAM | 4072 | 5 | 0 | 31 | |
| GSE74704 | EPCAM | 4072 | 9 | 1 | 10 | |
| TCGA | EPCAM | 4072 | 36 | 2 | 58 |
Total number of gains: 94; Total number of losses: 17; Total Number of normals: 377.
Somatic mutations of EPCAM:
Generating mutation plots.
Highly correlated genes for EPCAM:
Showing top 20/1443 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EPCAM | NCAPH | 0.761504 | 9 | 0 | 9 |
| EPCAM | RFC4 | 0.747237 | 13 | 0 | 12 |
| EPCAM | KIF14 | 0.737492 | 12 | 0 | 11 |
| EPCAM | SAPCD2 | 0.733641 | 6 | 0 | 6 |
| EPCAM | UBE2T | 0.73098 | 9 | 0 | 9 |
| EPCAM | RAD51AP1 | 0.727016 | 13 | 0 | 13 |
| EPCAM | MCM2 | 0.726849 | 13 | 0 | 12 |
| EPCAM | PSME4 | 0.724404 | 11 | 0 | 11 |
| EPCAM | ECT2 | 0.723049 | 13 | 0 | 12 |
| EPCAM | TPX2 | 0.720292 | 13 | 0 | 12 |
| EPCAM | CENPF | 0.718658 | 13 | 0 | 12 |
| EPCAM | TIMELESS | 0.716993 | 13 | 0 | 11 |
| EPCAM | USP14 | 0.714747 | 5 | 0 | 5 |
| EPCAM | TRIP13 | 0.714419 | 12 | 0 | 12 |
| EPCAM | TOP2A | 0.713761 | 13 | 0 | 13 |
| EPCAM | BUB1B | 0.712829 | 10 | 0 | 9 |
| EPCAM | FOXM1 | 0.71239 | 12 | 0 | 11 |
| EPCAM | PLK1 | 0.711834 | 10 | 0 | 9 |
| EPCAM | ACTL6A | 0.711723 | 13 | 0 | 13 |
| EPCAM | CDC45 | 0.711719 | 12 | 0 | 11 |
For details and further investigation, click here