| Full name: chloride channel accessory 1 | Alias Symbol: CaCC|CLCRG1 | ||
| Type: protein-coding gene | Cytoband: 1p22.3 | ||
| Entrez ID: 1179 | HGNC ID: HGNC:2015 | Ensembl Gene: ENSG00000016490 | OMIM ID: 603906 |
| Drug and gene relationship at DGIdb | |||
CLCA1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04972 | Pancreatic secretion |
Expression of CLCA1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLCA1 | 1179 | 210107_at | -0.0918 | 0.7616 | |
| GSE20347 | CLCA1 | 1179 | 210107_at | 0.0401 | 0.6367 | |
| GSE23400 | CLCA1 | 1179 | 210107_at | -0.0559 | 0.1206 | |
| GSE26886 | CLCA1 | 1179 | 210107_at | 0.0878 | 0.3178 | |
| GSE29001 | CLCA1 | 1179 | 210107_at | -0.0984 | 0.6151 | |
| GSE38129 | CLCA1 | 1179 | 210107_at | -0.0544 | 0.6218 | |
| GSE45670 | CLCA1 | 1179 | 210107_at | 0.0083 | 0.9446 | |
| GSE53622 | CLCA1 | 1179 | 105881 | -0.2833 | 0.0182 | |
| GSE53624 | CLCA1 | 1179 | 105881 | -0.1657 | 0.1492 | |
| GSE63941 | CLCA1 | 1179 | 210107_at | 0.0051 | 0.9801 | |
| GSE77861 | CLCA1 | 1179 | 210107_at | -0.0110 | 0.9439 |
Upregulated datasets: 0; Downregulated datasets: 0.
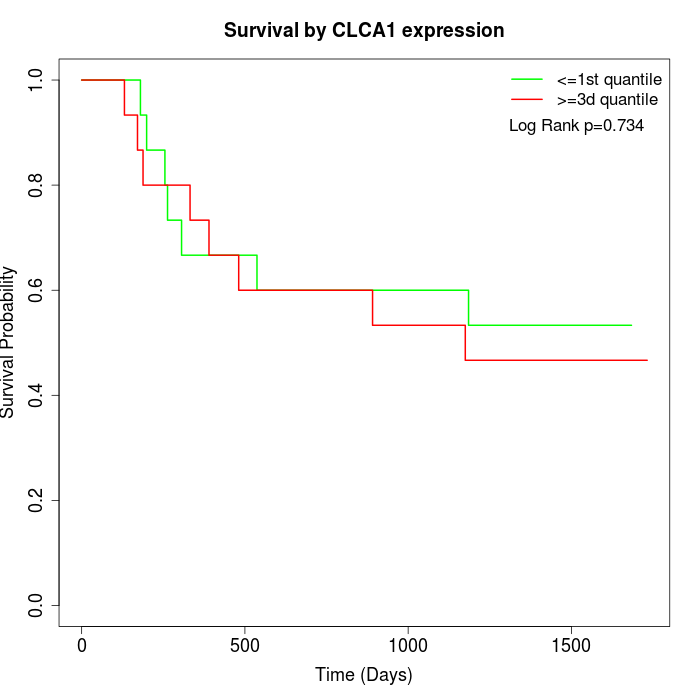
Survival by CLCA1 expression:
Note: Click image to view full size file.
Copy number change of CLCA1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLCA1 | 1179 | 0 | 8 | 22 | |
| GSE20123 | CLCA1 | 1179 | 0 | 8 | 22 | |
| GSE43470 | CLCA1 | 1179 | 2 | 2 | 39 | |
| GSE46452 | CLCA1 | 1179 | 1 | 1 | 57 | |
| GSE47630 | CLCA1 | 1179 | 8 | 5 | 27 | |
| GSE54993 | CLCA1 | 1179 | 0 | 1 | 69 | |
| GSE54994 | CLCA1 | 1179 | 6 | 3 | 44 | |
| GSE60625 | CLCA1 | 1179 | 0 | 0 | 11 | |
| GSE74703 | CLCA1 | 1179 | 1 | 2 | 33 | |
| GSE74704 | CLCA1 | 1179 | 0 | 5 | 15 | |
| TCGA | CLCA1 | 1179 | 6 | 23 | 67 |
Total number of gains: 24; Total number of losses: 58; Total Number of normals: 406.
Somatic mutations of CLCA1:
Generating mutation plots.
Highly correlated genes for CLCA1:
Showing top 20/759 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLCA1 | CNGB3 | 0.731019 | 4 | 0 | 4 |
| CLCA1 | CCDC30 | 0.719428 | 5 | 0 | 5 |
| CLCA1 | GLP1R | 0.715593 | 6 | 0 | 6 |
| CLCA1 | TNFSF8 | 0.712677 | 3 | 0 | 3 |
| CLCA1 | FASLG | 0.710382 | 4 | 0 | 4 |
| CLCA1 | NXPE4 | 0.706533 | 4 | 0 | 4 |
| CLCA1 | CYP11B2 | 0.704676 | 5 | 0 | 5 |
| CLCA1 | HNF4G | 0.699012 | 3 | 0 | 3 |
| CLCA1 | INSL5 | 0.697503 | 3 | 0 | 3 |
| CLCA1 | CA7 | 0.695987 | 5 | 0 | 5 |
| CLCA1 | KRTAP5-9 | 0.695439 | 4 | 0 | 4 |
| CLCA1 | KRT76 | 0.692797 | 5 | 0 | 5 |
| CLCA1 | SLC10A1 | 0.691336 | 5 | 0 | 5 |
| CLCA1 | GPLD1 | 0.690876 | 5 | 0 | 4 |
| CLCA1 | SMCP | 0.690574 | 5 | 0 | 5 |
| CLCA1 | ADCYAP1R1 | 0.689105 | 4 | 0 | 3 |
| CLCA1 | CIITA | 0.687766 | 7 | 0 | 7 |
| CLCA1 | ART1 | 0.686164 | 5 | 0 | 5 |
| CLCA1 | CST8 | 0.68584 | 3 | 0 | 3 |
| CLCA1 | KIR2DL2 | 0.682599 | 4 | 0 | 4 |
For details and further investigation, click here