| Full name: C-type lectin domain family 4 member D | Alias Symbol: Mpcl|CD368|MCL|Dectin-3 | ||
| Type: protein-coding gene | Cytoband: 12p13.31 | ||
| Entrez ID: 338339 | HGNC ID: HGNC:14554 | Ensembl Gene: ENSG00000166527 | OMIM ID: 609964 |
| Drug and gene relationship at DGIdb | |||
Expression of CLEC4D:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLEC4D | 338339 | 1552773_at | 0.0465 | 0.9280 | |
| GSE26886 | CLEC4D | 338339 | 1552773_at | -0.4355 | 0.0004 | |
| GSE45670 | CLEC4D | 338339 | 1552773_at | -0.0824 | 0.3029 | |
| GSE53622 | CLEC4D | 338339 | 101521 | 0.2126 | 0.0226 | |
| GSE53624 | CLEC4D | 338339 | 101521 | 0.3384 | 0.0003 | |
| GSE63941 | CLEC4D | 338339 | 1552772_at | 0.1196 | 0.3682 | |
| GSE77861 | CLEC4D | 338339 | 1552773_at | -0.0243 | 0.8465 | |
| GSE97050 | CLEC4D | 338339 | A_33_P3352578 | 0.0181 | 0.9373 | |
| TCGA | CLEC4D | 338339 | RNAseq | -0.5104 | 0.4348 |
Upregulated datasets: 0; Downregulated datasets: 0.
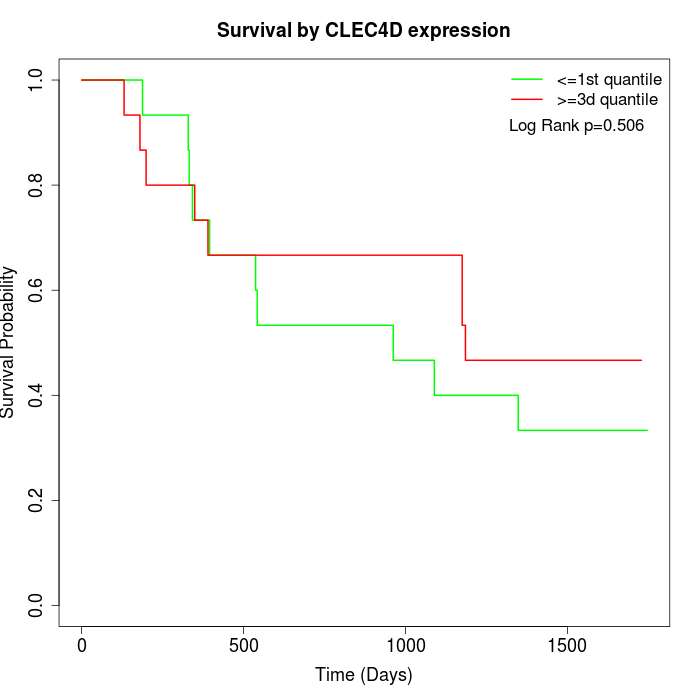
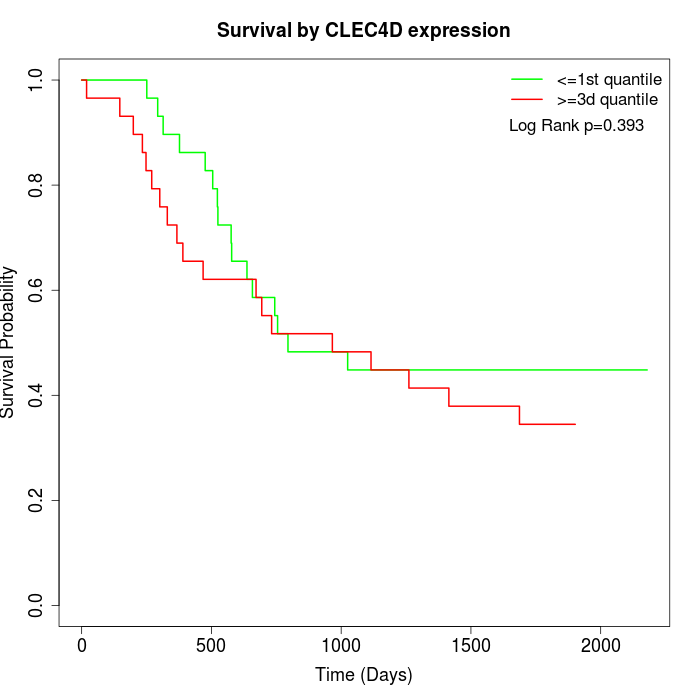
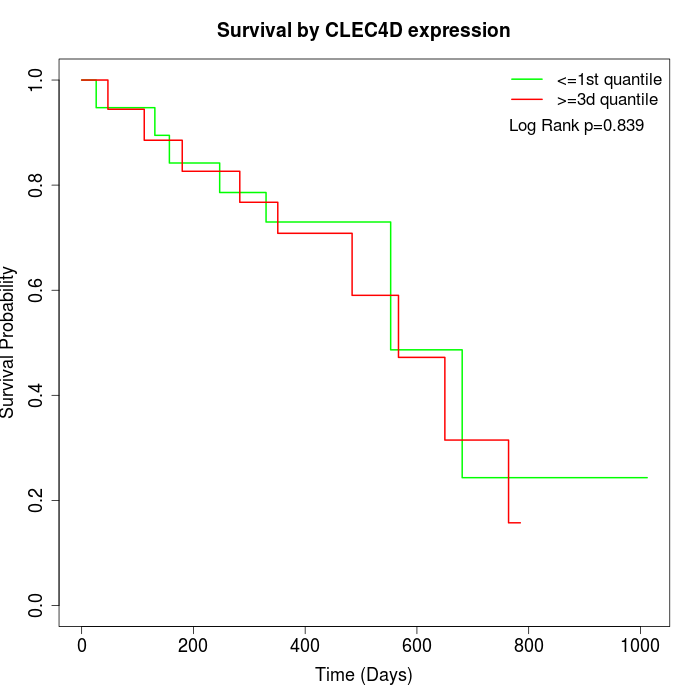
Survival by CLEC4D expression:
Note: Click image to view full size file.
Copy number change of CLEC4D:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLEC4D | 338339 | 6 | 3 | 21 | |
| GSE20123 | CLEC4D | 338339 | 6 | 3 | 21 | |
| GSE43470 | CLEC4D | 338339 | 7 | 3 | 33 | |
| GSE46452 | CLEC4D | 338339 | 10 | 1 | 48 | |
| GSE47630 | CLEC4D | 338339 | 12 | 2 | 26 | |
| GSE54993 | CLEC4D | 338339 | 1 | 10 | 59 | |
| GSE54994 | CLEC4D | 338339 | 9 | 2 | 42 | |
| GSE60625 | CLEC4D | 338339 | 0 | 1 | 10 | |
| GSE74703 | CLEC4D | 338339 | 7 | 2 | 27 | |
| GSE74704 | CLEC4D | 338339 | 4 | 2 | 14 | |
| TCGA | CLEC4D | 338339 | 40 | 6 | 50 |
Total number of gains: 102; Total number of losses: 35; Total Number of normals: 351.
Somatic mutations of CLEC4D:
Generating mutation plots.
Highly correlated genes for CLEC4D:
Showing top 20/34 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLEC4D | TSSK3 | 0.791519 | 3 | 0 | 3 |
| CLEC4D | HTR3A | 0.753941 | 3 | 0 | 3 |
| CLEC4D | UGT3A1 | 0.742782 | 3 | 0 | 3 |
| CLEC4D | GRM6 | 0.710808 | 3 | 0 | 3 |
| CLEC4D | KRTAP4-8 | 0.708667 | 3 | 0 | 3 |
| CLEC4D | ATP2B3 | 0.696058 | 3 | 0 | 3 |
| CLEC4D | CHTF8 | 0.686381 | 3 | 0 | 3 |
| CLEC4D | BRAF | 0.685018 | 3 | 0 | 3 |
| CLEC4D | OR12D3 | 0.659812 | 4 | 0 | 4 |
| CLEC4D | SPEF2 | 0.659669 | 3 | 0 | 3 |
| CLEC4D | EPOR | 0.656708 | 3 | 0 | 3 |
| CLEC4D | ONECUT1 | 0.649247 | 4 | 0 | 4 |
| CLEC4D | GPR52 | 0.645865 | 3 | 0 | 3 |
| CLEC4D | TM6SF1 | 0.643647 | 3 | 0 | 3 |
| CLEC4D | CCL17 | 0.636729 | 3 | 0 | 3 |
| CLEC4D | PRAMEF12 | 0.619178 | 6 | 0 | 3 |
| CLEC4D | ST8SIA3 | 0.615267 | 4 | 0 | 3 |
| CLEC4D | ZNF236 | 0.605764 | 4 | 0 | 3 |
| CLEC4D | FCGR2B | 0.599606 | 3 | 0 | 3 |
| CLEC4D | FPR1 | 0.596809 | 5 | 0 | 4 |
For details and further investigation, click here