| Full name: chondroitin sulfate N-acetylgalactosaminyltransferase 2 | Alias Symbol: GALNACT2|MGC40204|PRO0082|GALNACT-2 | ||
| Type: protein-coding gene | Cytoband: 10q11.21 | ||
| Entrez ID: 55454 | HGNC ID: HGNC:24292 | Ensembl Gene: ENSG00000169826 | OMIM ID: 616616 |
| Drug and gene relationship at DGIdb | |||
Expression of CSGALNACT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CSGALNACT2 | 55454 | 218871_x_at | 0.3242 | 0.5189 | |
| GSE20347 | CSGALNACT2 | 55454 | 218871_x_at | 1.0224 | 0.0000 | |
| GSE23400 | CSGALNACT2 | 55454 | 222235_s_at | 0.6667 | 0.0000 | |
| GSE26886 | CSGALNACT2 | 55454 | 218871_x_at | 1.6636 | 0.0000 | |
| GSE29001 | CSGALNACT2 | 55454 | 218871_x_at | 0.8601 | 0.0081 | |
| GSE38129 | CSGALNACT2 | 55454 | 218871_x_at | 0.7587 | 0.0001 | |
| GSE45670 | CSGALNACT2 | 55454 | 218871_x_at | 0.0417 | 0.9119 | |
| GSE53622 | CSGALNACT2 | 55454 | 35129 | 1.0110 | 0.0000 | |
| GSE53624 | CSGALNACT2 | 55454 | 35129 | 0.9543 | 0.0000 | |
| GSE63941 | CSGALNACT2 | 55454 | 218871_x_at | -2.3771 | 0.0007 | |
| GSE77861 | CSGALNACT2 | 55454 | 218871_x_at | 0.9666 | 0.0731 | |
| GSE97050 | CSGALNACT2 | 55454 | A_23_P149892 | 0.5949 | 0.1581 | |
| SRP007169 | CSGALNACT2 | 55454 | RNAseq | 2.7431 | 0.0000 | |
| SRP008496 | CSGALNACT2 | 55454 | RNAseq | 2.7395 | 0.0000 | |
| SRP064894 | CSGALNACT2 | 55454 | RNAseq | 0.7442 | 0.0411 | |
| SRP133303 | CSGALNACT2 | 55454 | RNAseq | 1.2281 | 0.0000 | |
| SRP159526 | CSGALNACT2 | 55454 | RNAseq | 0.4222 | 0.1804 | |
| SRP193095 | CSGALNACT2 | 55454 | RNAseq | 1.0672 | 0.0000 | |
| SRP219564 | CSGALNACT2 | 55454 | RNAseq | 0.4356 | 0.2482 | |
| TCGA | CSGALNACT2 | 55454 | RNAseq | 0.1645 | 0.0294 |
Upregulated datasets: 7; Downregulated datasets: 1.
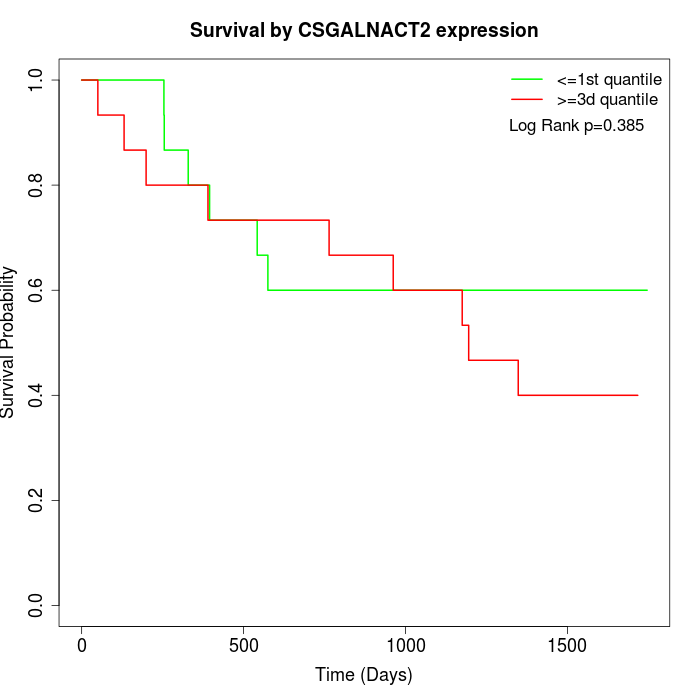
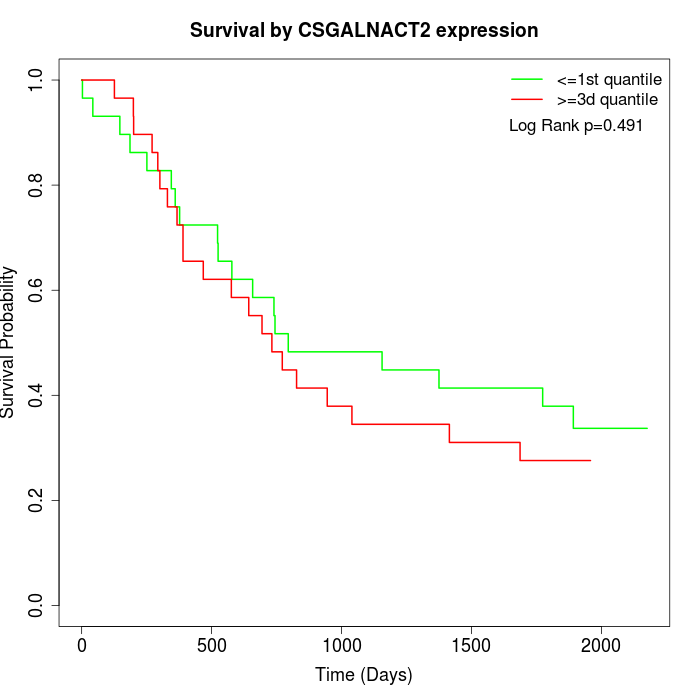
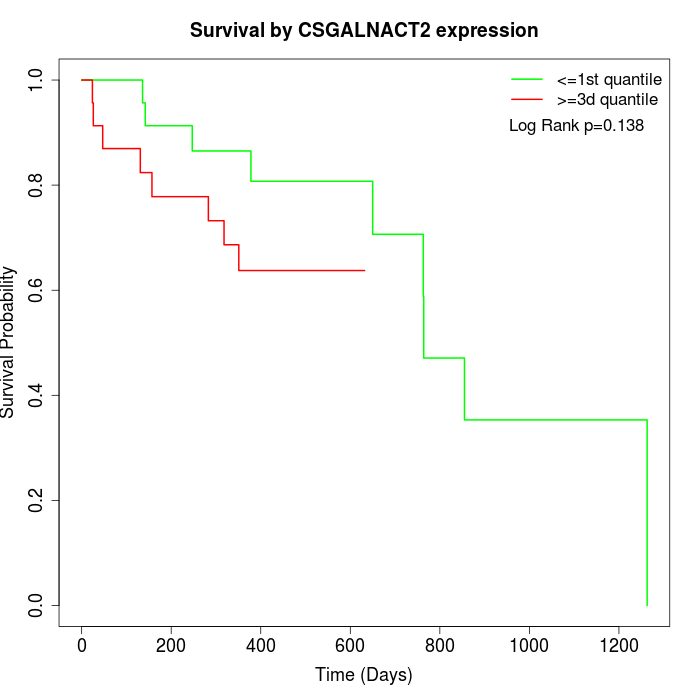
Survival by CSGALNACT2 expression:
Note: Click image to view full size file.
Copy number change of CSGALNACT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CSGALNACT2 | 55454 | 2 | 6 | 22 | |
| GSE20123 | CSGALNACT2 | 55454 | 3 | 5 | 22 | |
| GSE43470 | CSGALNACT2 | 55454 | 1 | 6 | 36 | |
| GSE46452 | CSGALNACT2 | 55454 | 0 | 13 | 46 | |
| GSE47630 | CSGALNACT2 | 55454 | 4 | 12 | 24 | |
| GSE54993 | CSGALNACT2 | 55454 | 9 | 0 | 61 | |
| GSE54994 | CSGALNACT2 | 55454 | 2 | 10 | 41 | |
| GSE60625 | CSGALNACT2 | 55454 | 0 | 0 | 11 | |
| GSE74703 | CSGALNACT2 | 55454 | 1 | 3 | 32 | |
| GSE74704 | CSGALNACT2 | 55454 | 2 | 4 | 14 | |
| TCGA | CSGALNACT2 | 55454 | 16 | 20 | 60 |
Total number of gains: 40; Total number of losses: 79; Total Number of normals: 369.
Somatic mutations of CSGALNACT2:
Generating mutation plots.
Highly correlated genes for CSGALNACT2:
Showing top 20/408 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CSGALNACT2 | COL5A2 | 0.767933 | 11 | 0 | 11 |
| CSGALNACT2 | COL6A3 | 0.765338 | 11 | 0 | 11 |
| CSGALNACT2 | TMEM263 | 0.760491 | 7 | 0 | 7 |
| CSGALNACT2 | COL5A1 | 0.748444 | 11 | 0 | 11 |
| CSGALNACT2 | LHFPL2 | 0.741148 | 12 | 0 | 12 |
| CSGALNACT2 | VCAN | 0.734587 | 12 | 0 | 11 |
| CSGALNACT2 | DACT1 | 0.733491 | 11 | 0 | 11 |
| CSGALNACT2 | CHSY1 | 0.727052 | 11 | 0 | 11 |
| CSGALNACT2 | NID2 | 0.719568 | 11 | 0 | 10 |
| CSGALNACT2 | PMEPA1 | 0.717328 | 11 | 0 | 11 |
| CSGALNACT2 | SMIM3 | 0.712495 | 7 | 0 | 7 |
| CSGALNACT2 | FAP | 0.710911 | 10 | 0 | 9 |
| CSGALNACT2 | OLFML2B | 0.709872 | 11 | 0 | 11 |
| CSGALNACT2 | NOX4 | 0.708098 | 11 | 0 | 11 |
| CSGALNACT2 | LUM | 0.70657 | 12 | 0 | 10 |
| CSGALNACT2 | ADAMTS6 | 0.702122 | 5 | 0 | 5 |
| CSGALNACT2 | MFAP2 | 0.701539 | 10 | 0 | 10 |
| CSGALNACT2 | INHBA | 0.701002 | 12 | 0 | 11 |
| CSGALNACT2 | CTSL | 0.697387 | 11 | 0 | 10 |
| CSGALNACT2 | TGFBI | 0.69574 | 10 | 0 | 9 |
For details and further investigation, click here