| Full name: fibroblast activation protein alpha | Alias Symbol: DPPIV | ||
| Type: protein-coding gene | Cytoband: 2q24.2 | ||
| Entrez ID: 2191 | HGNC ID: HGNC:3590 | Ensembl Gene: ENSG00000078098 | OMIM ID: 600403 |
| Drug and gene relationship at DGIdb | |||
Expression of FAP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FAP | 2191 | 209955_s_at | 1.4827 | 0.1173 | |
| GSE20347 | FAP | 2191 | 209955_s_at | 1.2921 | 0.0000 | |
| GSE23400 | FAP | 2191 | 209955_s_at | 1.2914 | 0.0000 | |
| GSE26886 | FAP | 2191 | 209955_s_at | 1.1404 | 0.0036 | |
| GSE29001 | FAP | 2191 | 209955_s_at | 0.7917 | 0.0034 | |
| GSE38129 | FAP | 2191 | 209955_s_at | 1.7507 | 0.0000 | |
| GSE45670 | FAP | 2191 | 209955_s_at | 0.5508 | 0.4423 | |
| GSE63941 | FAP | 2191 | 209955_s_at | -6.5512 | 0.0000 | |
| GSE77861 | FAP | 2191 | 209955_s_at | 0.3241 | 0.0488 | |
| GSE97050 | FAP | 2191 | A_23_P56746 | 1.1869 | 0.1156 | |
| SRP007169 | FAP | 2191 | RNAseq | 5.2633 | 0.0000 | |
| SRP008496 | FAP | 2191 | RNAseq | 6.4952 | 0.0000 | |
| SRP064894 | FAP | 2191 | RNAseq | 3.4610 | 0.0000 | |
| SRP133303 | FAP | 2191 | RNAseq | 3.5682 | 0.0000 | |
| SRP159526 | FAP | 2191 | RNAseq | 2.6269 | 0.0000 | |
| SRP219564 | FAP | 2191 | RNAseq | 1.8226 | 0.0468 | |
| TCGA | FAP | 2191 | RNAseq | 1.4669 | 0.0000 |
Upregulated datasets: 11; Downregulated datasets: 1.
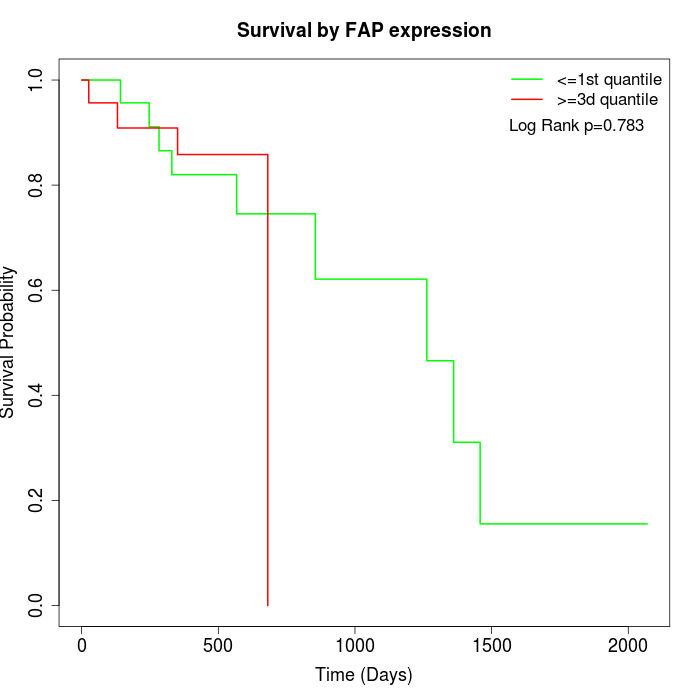
Survival by FAP expression:
Note: Click image to view full size file.
Copy number change of FAP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FAP | 2191 | 4 | 1 | 25 | |
| GSE20123 | FAP | 2191 | 4 | 1 | 25 | |
| GSE43470 | FAP | 2191 | 4 | 1 | 38 | |
| GSE46452 | FAP | 2191 | 1 | 4 | 54 | |
| GSE47630 | FAP | 2191 | 5 | 3 | 32 | |
| GSE54993 | FAP | 2191 | 0 | 5 | 65 | |
| GSE54994 | FAP | 2191 | 11 | 2 | 40 | |
| GSE60625 | FAP | 2191 | 0 | 3 | 8 | |
| GSE74703 | FAP | 2191 | 3 | 1 | 32 | |
| GSE74704 | FAP | 2191 | 3 | 0 | 17 | |
| TCGA | FAP | 2191 | 23 | 10 | 63 |
Total number of gains: 58; Total number of losses: 31; Total Number of normals: 399.
Somatic mutations of FAP:
Generating mutation plots.
Highly correlated genes for FAP:
Showing top 20/996 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FAP | COL5A2 | 0.85175 | 11 | 0 | 11 |
| FAP | COL5A1 | 0.844199 | 11 | 0 | 11 |
| FAP | COL1A2 | 0.840917 | 11 | 0 | 11 |
| FAP | COL6A3 | 0.837233 | 11 | 0 | 11 |
| FAP | POSTN | 0.834793 | 11 | 0 | 11 |
| FAP | VCAN | 0.833499 | 11 | 0 | 11 |
| FAP | INHBA | 0.83022 | 10 | 0 | 10 |
| FAP | ADAMTS12 | 0.821195 | 6 | 0 | 6 |
| FAP | SPARC | 0.820787 | 11 | 0 | 10 |
| FAP | COL1A1 | 0.819773 | 10 | 0 | 10 |
| FAP | CTSK | 0.813701 | 11 | 0 | 11 |
| FAP | COL3A1 | 0.810945 | 11 | 0 | 11 |
| FAP | OLFML2B | 0.809546 | 11 | 0 | 11 |
| FAP | BGN | 0.808817 | 11 | 0 | 11 |
| FAP | LOXL2 | 0.808272 | 10 | 0 | 10 |
| FAP | THY1 | 0.808022 | 11 | 0 | 11 |
| FAP | NID2 | 0.806339 | 10 | 0 | 10 |
| FAP | NOX4 | 0.799318 | 11 | 0 | 11 |
| FAP | SULF1 | 0.798959 | 11 | 0 | 11 |
| FAP | CDH11 | 0.797213 | 11 | 0 | 11 |
For details and further investigation, click here