| Full name: C-X-C motif chemokine receptor 4 | Alias Symbol: LESTR|NPY3R|HM89|NPYY3R|D2S201E|fusin|HSY3RR|NPYR|CD184 | ||
| Type: protein-coding gene | Cytoband: 2q22.1 | ||
| Entrez ID: 7852 | HGNC ID: HGNC:2561 | Ensembl Gene: ENSG00000121966 | OMIM ID: 162643 |
| Drug and gene relationship at DGIdb | |||
CXCR4 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway | |
| hsa04360 | Axon guidance | |
| hsa04670 | Leukocyte transendothelial migration |
Expression of CXCR4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CXCR4 | 7852 | 217028_at | 0.3000 | 0.8568 | |
| GSE20347 | CXCR4 | 7852 | 217028_at | 0.7368 | 0.1149 | |
| GSE23400 | CXCR4 | 7852 | 211919_s_at | 0.3381 | 0.0149 | |
| GSE26886 | CXCR4 | 7852 | 217028_at | 2.2403 | 0.0005 | |
| GSE29001 | CXCR4 | 7852 | 217028_at | 1.2173 | 0.0580 | |
| GSE38129 | CXCR4 | 7852 | 217028_at | 0.6775 | 0.1048 | |
| GSE45670 | CXCR4 | 7852 | 217028_at | -0.5756 | 0.3314 | |
| GSE53622 | CXCR4 | 7852 | 82504 | 0.1458 | 0.4015 | |
| GSE53624 | CXCR4 | 7852 | 82504 | -0.2530 | 0.0540 | |
| GSE63941 | CXCR4 | 7852 | 209201_x_at | 0.6206 | 0.5032 | |
| GSE77861 | CXCR4 | 7852 | 211919_s_at | 0.4831 | 0.1647 | |
| GSE97050 | CXCR4 | 7852 | A_23_P102000 | 0.9024 | 0.2209 | |
| SRP007169 | CXCR4 | 7852 | RNAseq | 1.9790 | 0.0104 | |
| SRP008496 | CXCR4 | 7852 | RNAseq | 1.1460 | 0.0216 | |
| SRP064894 | CXCR4 | 7852 | RNAseq | 0.9161 | 0.0105 | |
| SRP133303 | CXCR4 | 7852 | RNAseq | 0.5139 | 0.1849 | |
| SRP159526 | CXCR4 | 7852 | RNAseq | -0.1341 | 0.8517 | |
| SRP193095 | CXCR4 | 7852 | RNAseq | 0.8785 | 0.0172 | |
| SRP219564 | CXCR4 | 7852 | RNAseq | 1.1230 | 0.0131 | |
| TCGA | CXCR4 | 7852 | RNAseq | -0.1003 | 0.4207 |
Upregulated datasets: 4; Downregulated datasets: 0.
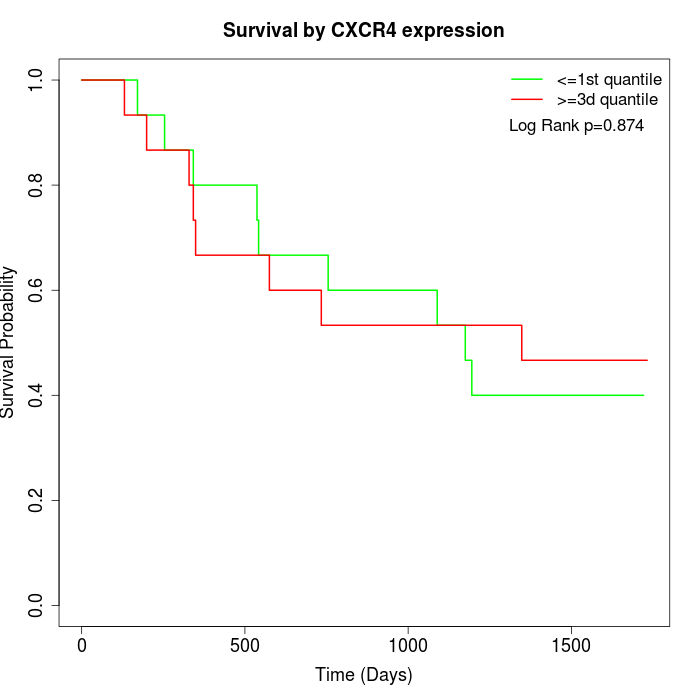
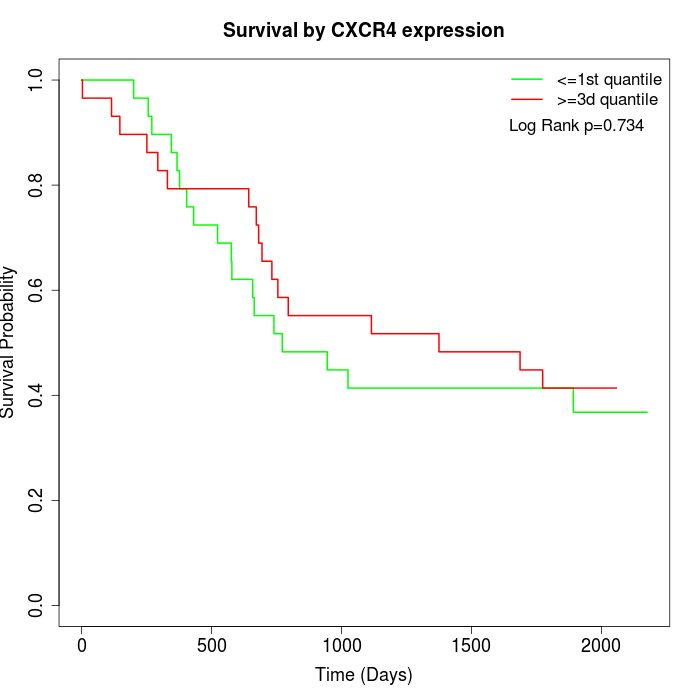
Survival by CXCR4 expression:
Note: Click image to view full size file.
Copy number change of CXCR4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CXCR4 | 7852 | 2 | 4 | 24 | |
| GSE20123 | CXCR4 | 7852 | 2 | 4 | 24 | |
| GSE43470 | CXCR4 | 7852 | 2 | 1 | 40 | |
| GSE46452 | CXCR4 | 7852 | 1 | 4 | 54 | |
| GSE47630 | CXCR4 | 7852 | 5 | 2 | 33 | |
| GSE54993 | CXCR4 | 7852 | 0 | 6 | 64 | |
| GSE54994 | CXCR4 | 7852 | 9 | 0 | 44 | |
| GSE60625 | CXCR4 | 7852 | 0 | 3 | 8 | |
| GSE74703 | CXCR4 | 7852 | 2 | 1 | 33 | |
| GSE74704 | CXCR4 | 7852 | 1 | 1 | 18 | |
| TCGA | CXCR4 | 7852 | 21 | 10 | 65 |
Total number of gains: 45; Total number of losses: 36; Total Number of normals: 407.
Somatic mutations of CXCR4:
Generating mutation plots.
Highly correlated genes for CXCR4:
Showing top 20/584 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CXCR4 | PSME4 | 0.760253 | 3 | 0 | 3 |
| CXCR4 | CXCL3 | 0.745922 | 3 | 0 | 3 |
| CXCR4 | OCIAD2 | 0.726731 | 3 | 0 | 3 |
| CXCR4 | SLITRK6 | 0.714913 | 3 | 0 | 3 |
| CXCR4 | UBE2T | 0.71449 | 3 | 0 | 3 |
| CXCR4 | DHCR7 | 0.713177 | 3 | 0 | 3 |
| CXCR4 | FMNL2 | 0.708878 | 3 | 0 | 3 |
| CXCR4 | PARVG | 0.706117 | 6 | 0 | 5 |
| CXCR4 | EME1 | 0.705303 | 3 | 0 | 3 |
| CXCR4 | PTK7 | 0.702636 | 3 | 0 | 3 |
| CXCR4 | CD53 | 0.700064 | 8 | 0 | 7 |
| CXCR4 | LOXL2 | 0.697936 | 3 | 0 | 3 |
| CXCR4 | CD226 | 0.692282 | 4 | 0 | 4 |
| CXCR4 | ST6GAL1 | 0.69189 | 12 | 0 | 12 |
| CXCR4 | ERVMER34-1 | 0.690606 | 4 | 0 | 3 |
| CXCR4 | TOP1MT | 0.686056 | 3 | 0 | 3 |
| CXCR4 | ECE1 | 0.684891 | 3 | 0 | 3 |
| CXCR4 | GPR158 | 0.680124 | 3 | 0 | 3 |
| CXCR4 | P4HA1 | 0.679081 | 4 | 0 | 3 |
| CXCR4 | TSPAN1 | 0.678439 | 3 | 0 | 3 |
For details and further investigation, click here