| Full name: early growth response 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10q21.3 | ||
| Entrez ID: 1959 | HGNC ID: HGNC:3239 | Ensembl Gene: ENSG00000122877 | OMIM ID: 129010 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
EGR2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05161 | Hepatitis B |
Expression of EGR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EGR2 | 1959 | 205249_at | 0.8425 | 0.2805 | |
| GSE20347 | EGR2 | 1959 | 205249_at | 0.7129 | 0.0162 | |
| GSE23400 | EGR2 | 1959 | 205249_at | 0.2612 | 0.0158 | |
| GSE26886 | EGR2 | 1959 | 205249_at | 0.6905 | 0.0296 | |
| GSE29001 | EGR2 | 1959 | 205249_at | 0.5631 | 0.1534 | |
| GSE38129 | EGR2 | 1959 | 205249_at | 0.7148 | 0.0167 | |
| GSE45670 | EGR2 | 1959 | 205249_at | 0.0837 | 0.8929 | |
| GSE53622 | EGR2 | 1959 | 14756 | 0.5732 | 0.0097 | |
| GSE53624 | EGR2 | 1959 | 14756 | 1.2476 | 0.0000 | |
| GSE63941 | EGR2 | 1959 | 205249_at | -1.2443 | 0.3230 | |
| GSE77861 | EGR2 | 1959 | 205249_at | 0.8317 | 0.0353 | |
| GSE97050 | EGR2 | 1959 | A_23_P46936 | -0.0246 | 0.9774 | |
| SRP007169 | EGR2 | 1959 | RNAseq | 2.6717 | 0.0114 | |
| SRP008496 | EGR2 | 1959 | RNAseq | 1.6687 | 0.0022 | |
| SRP064894 | EGR2 | 1959 | RNAseq | 1.4119 | 0.0011 | |
| SRP133303 | EGR2 | 1959 | RNAseq | 1.1917 | 0.0034 | |
| SRP159526 | EGR2 | 1959 | RNAseq | 1.9483 | 0.0000 | |
| SRP193095 | EGR2 | 1959 | RNAseq | 0.9912 | 0.0653 | |
| SRP219564 | EGR2 | 1959 | RNAseq | 1.2649 | 0.2441 | |
| TCGA | EGR2 | 1959 | RNAseq | 0.4254 | 0.0245 |
Upregulated datasets: 6; Downregulated datasets: 0.
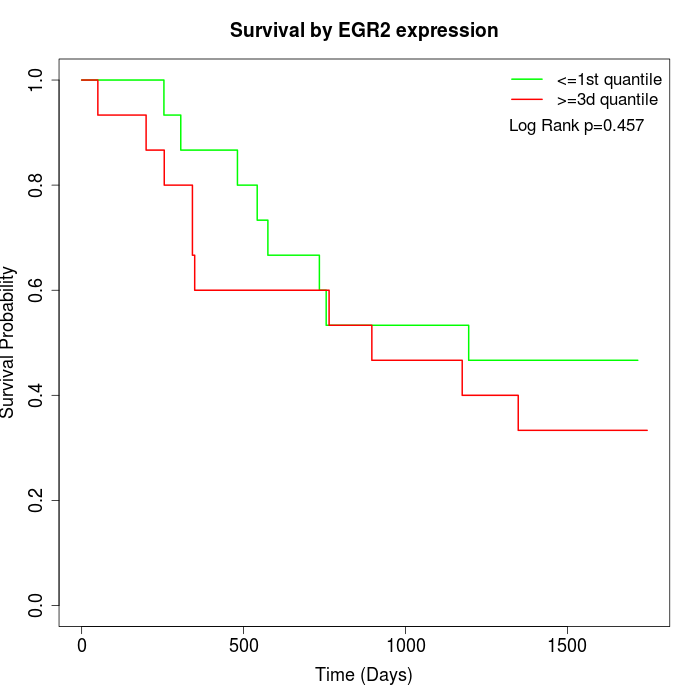
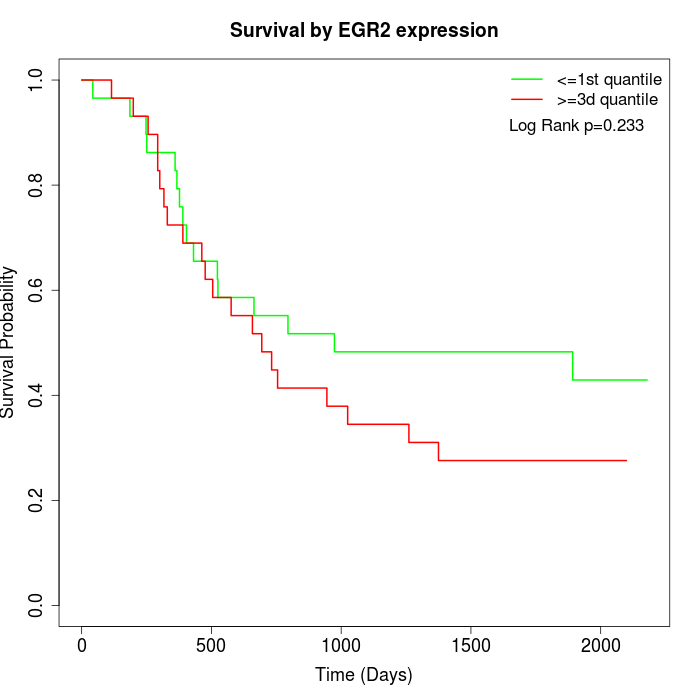
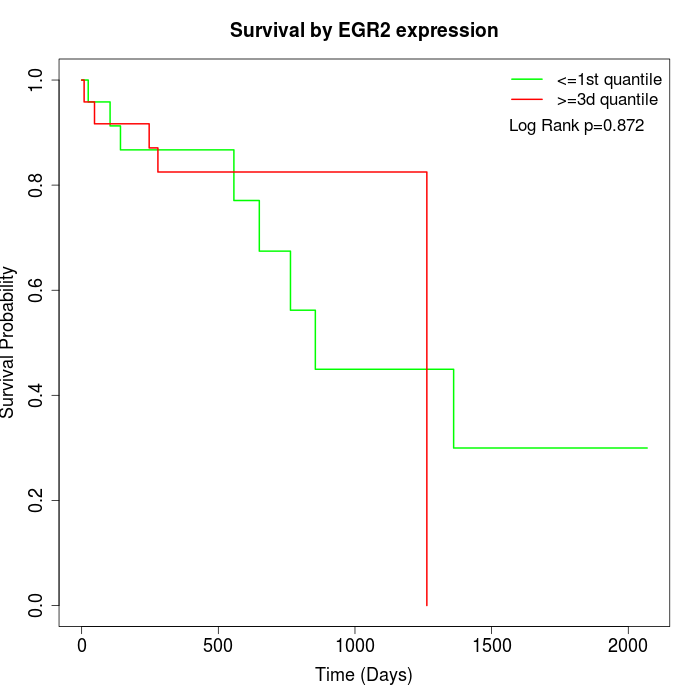
Survival by EGR2 expression:
Note: Click image to view full size file.
Copy number change of EGR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EGR2 | 1959 | 2 | 7 | 21 | |
| GSE20123 | EGR2 | 1959 | 2 | 7 | 21 | |
| GSE43470 | EGR2 | 1959 | 2 | 5 | 36 | |
| GSE46452 | EGR2 | 1959 | 0 | 11 | 48 | |
| GSE47630 | EGR2 | 1959 | 4 | 12 | 24 | |
| GSE54993 | EGR2 | 1959 | 9 | 0 | 61 | |
| GSE54994 | EGR2 | 1959 | 4 | 9 | 40 | |
| GSE60625 | EGR2 | 1959 | 0 | 0 | 11 | |
| GSE74703 | EGR2 | 1959 | 2 | 3 | 31 | |
| GSE74704 | EGR2 | 1959 | 1 | 5 | 14 | |
| TCGA | EGR2 | 1959 | 12 | 22 | 62 |
Total number of gains: 38; Total number of losses: 81; Total Number of normals: 369.
Somatic mutations of EGR2:
Generating mutation plots.
Highly correlated genes for EGR2:
Showing top 20/521 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EGR2 | REC8 | 0.732346 | 3 | 0 | 3 |
| EGR2 | C1orf115 | 0.710075 | 3 | 0 | 3 |
| EGR2 | EGR3 | 0.709235 | 11 | 0 | 10 |
| EGR2 | TRIM47 | 0.701357 | 3 | 0 | 3 |
| EGR2 | ADAMTS15 | 0.701 | 3 | 0 | 3 |
| EGR2 | CTF1 | 0.695145 | 3 | 0 | 3 |
| EGR2 | IFT172 | 0.693189 | 3 | 0 | 3 |
| EGR2 | CCDC127 | 0.685705 | 3 | 0 | 3 |
| EGR2 | MEX3B | 0.679011 | 5 | 0 | 4 |
| EGR2 | FGF1 | 0.676169 | 3 | 0 | 3 |
| EGR2 | PLEK | 0.670768 | 3 | 0 | 3 |
| EGR2 | CXCL5 | 0.667883 | 4 | 0 | 3 |
| EGR2 | PTGES2 | 0.667066 | 3 | 0 | 3 |
| EGR2 | GCLC | 0.664846 | 4 | 0 | 3 |
| EGR2 | DOCK11 | 0.661493 | 4 | 0 | 3 |
| EGR2 | FANCC | 0.6593 | 4 | 0 | 3 |
| EGR2 | CERK | 0.658716 | 3 | 0 | 3 |
| EGR2 | WFS1 | 0.656176 | 4 | 0 | 3 |
| EGR2 | SLC25A12 | 0.654784 | 3 | 0 | 3 |
| EGR2 | PIM2 | 0.654528 | 3 | 0 | 3 |
For details and further investigation, click here