| Full name: serine protease 22 | Alias Symbol: hBSSP-4|BSSP-4|SP001LA | ||
| Type: protein-coding gene | Cytoband: 16p13.3 | ||
| Entrez ID: 64063 | HGNC ID: HGNC:14368 | Ensembl Gene: ENSG00000005001 | OMIM ID: 609343 |
| Drug and gene relationship at DGIdb | |||
Expression of PRSS22:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRSS22 | 64063 | 205847_at | -0.5229 | 0.5699 | |
| GSE20347 | PRSS22 | 64063 | 205847_at | -0.6935 | 0.0001 | |
| GSE23400 | PRSS22 | 64063 | 205847_at | -1.0948 | 0.0000 | |
| GSE26886 | PRSS22 | 64063 | 205847_at | -1.3029 | 0.0001 | |
| GSE29001 | PRSS22 | 64063 | 205847_at | -0.5486 | 0.0098 | |
| GSE38129 | PRSS22 | 64063 | 205847_at | -0.5046 | 0.0062 | |
| GSE45670 | PRSS22 | 64063 | 205847_at | 0.3352 | 0.0549 | |
| GSE53622 | PRSS22 | 64063 | 19440 | -0.8042 | 0.0053 | |
| GSE53624 | PRSS22 | 64063 | 19440 | -1.1429 | 0.0000 | |
| GSE63941 | PRSS22 | 64063 | 205847_at | 1.0287 | 0.0319 | |
| GSE77861 | PRSS22 | 64063 | 205847_at | -0.6198 | 0.0175 | |
| GSE97050 | PRSS22 | 64063 | A_23_P400298 | 0.3285 | 0.6756 | |
| SRP007169 | PRSS22 | 64063 | RNAseq | -2.9864 | 0.0000 | |
| SRP008496 | PRSS22 | 64063 | RNAseq | -3.5000 | 0.0000 | |
| SRP064894 | PRSS22 | 64063 | RNAseq | -2.1285 | 0.0000 | |
| SRP133303 | PRSS22 | 64063 | RNAseq | -0.9418 | 0.0250 | |
| SRP159526 | PRSS22 | 64063 | RNAseq | -2.8470 | 0.0000 | |
| SRP193095 | PRSS22 | 64063 | RNAseq | -2.1288 | 0.0000 | |
| SRP219564 | PRSS22 | 64063 | RNAseq | -1.4040 | 0.2620 | |
| TCGA | PRSS22 | 64063 | RNAseq | 0.5725 | 0.0306 |
Upregulated datasets: 1; Downregulated datasets: 8.
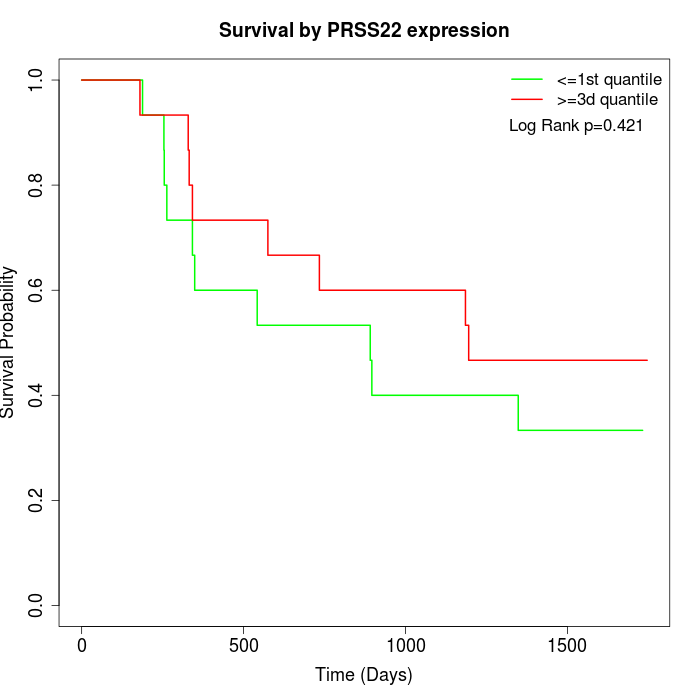
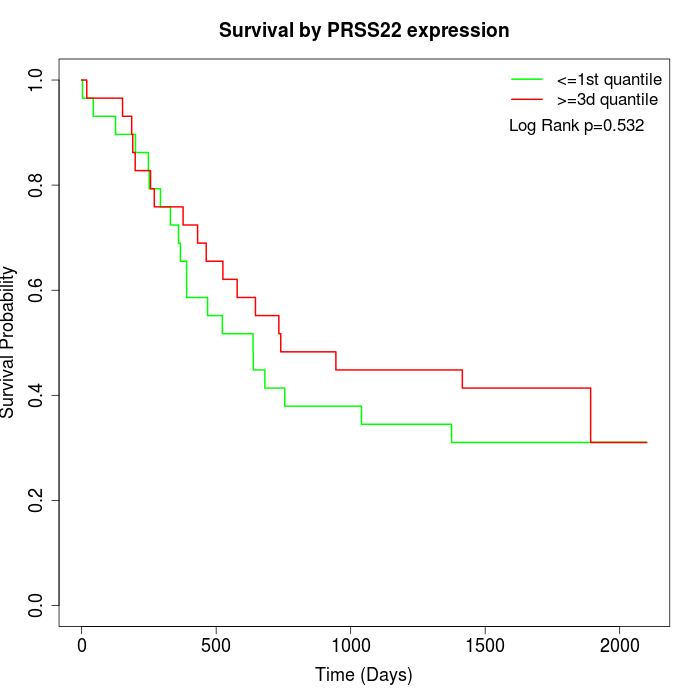
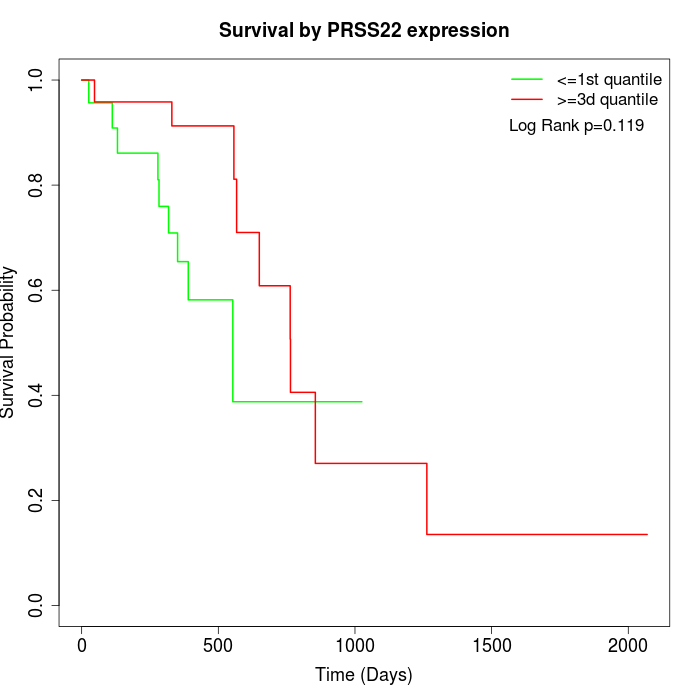
Survival by PRSS22 expression:
Note: Click image to view full size file.
Copy number change of PRSS22:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PRSS22 | 64063 | 5 | 5 | 20 | |
| GSE20123 | PRSS22 | 64063 | 5 | 4 | 21 | |
| GSE43470 | PRSS22 | 64063 | 4 | 7 | 32 | |
| GSE46452 | PRSS22 | 64063 | 38 | 1 | 20 | |
| GSE47630 | PRSS22 | 64063 | 13 | 6 | 21 | |
| GSE54993 | PRSS22 | 64063 | 3 | 5 | 62 | |
| GSE54994 | PRSS22 | 64063 | 5 | 9 | 39 | |
| GSE60625 | PRSS22 | 64063 | 4 | 0 | 7 | |
| GSE74703 | PRSS22 | 64063 | 4 | 5 | 27 | |
| GSE74704 | PRSS22 | 64063 | 3 | 2 | 15 | |
| TCGA | PRSS22 | 64063 | 19 | 13 | 64 |
Total number of gains: 103; Total number of losses: 57; Total Number of normals: 328.
Somatic mutations of PRSS22:
Generating mutation plots.
Highly correlated genes for PRSS22:
Showing top 20/1164 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRSS22 | EPHA2 | 0.752986 | 12 | 0 | 12 |
| PRSS22 | TMEM40 | 0.737573 | 12 | 0 | 12 |
| PRSS22 | PLIN3 | 0.734553 | 11 | 0 | 10 |
| PRSS22 | CLIC3 | 0.726789 | 9 | 0 | 9 |
| PRSS22 | NUAK2 | 0.72429 | 4 | 0 | 4 |
| PRSS22 | EPS8L1 | 0.718149 | 12 | 0 | 12 |
| PRSS22 | IL36A | 0.717804 | 9 | 0 | 9 |
| PRSS22 | SH3GL1 | 0.717139 | 10 | 0 | 10 |
| PRSS22 | TGM1 | 0.7171 | 12 | 0 | 11 |
| PRSS22 | NIPAL1 | 0.714368 | 3 | 0 | 3 |
| PRSS22 | TMEM234 | 0.713144 | 3 | 0 | 3 |
| PRSS22 | PRSS27 | 0.712508 | 7 | 0 | 7 |
| PRSS22 | EPS8L2 | 0.710014 | 10 | 0 | 10 |
| PRSS22 | EPN3 | 0.709638 | 11 | 0 | 10 |
| PRSS22 | PADI1 | 0.70822 | 8 | 0 | 8 |
| PRSS22 | PSCA | 0.707301 | 8 | 0 | 8 |
| PRSS22 | RAB25 | 0.705602 | 12 | 0 | 11 |
| PRSS22 | CEACAM1 | 0.700772 | 10 | 0 | 10 |
| PRSS22 | TTC9 | 0.699564 | 11 | 0 | 11 |
| PRSS22 | ARHGAP27 | 0.696856 | 8 | 0 | 8 |
For details and further investigation, click here