| Full name: family with sequence similarity 50 member A | Alias Symbol: DXS9928E|XAP5|HXC-26|9F | ||
| Type: protein-coding gene | Cytoband: Xq28 | ||
| Entrez ID: 9130 | HGNC ID: HGNC:18786 | Ensembl Gene: ENSG00000071859 | OMIM ID: 300453 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of FAM50A:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FAM50A | 9130 | 203262_s_at | -0.1103 | 0.8506 | |
| GSE20347 | FAM50A | 9130 | 203262_s_at | 0.2823 | 0.1052 | |
| GSE23400 | FAM50A | 9130 | 203262_s_at | 0.3873 | 0.0000 | |
| GSE26886 | FAM50A | 9130 | 203262_s_at | 0.6915 | 0.0017 | |
| GSE29001 | FAM50A | 9130 | 203262_s_at | 0.2247 | 0.6758 | |
| GSE38129 | FAM50A | 9130 | 203262_s_at | 0.1776 | 0.3722 | |
| GSE45670 | FAM50A | 9130 | 203262_s_at | 0.3918 | 0.0439 | |
| GSE53622 | FAM50A | 9130 | 57507 | 0.2840 | 0.0000 | |
| GSE53624 | FAM50A | 9130 | 57507 | 0.3860 | 0.0000 | |
| GSE63941 | FAM50A | 9130 | 203262_s_at | -1.3436 | 0.1297 | |
| GSE77861 | FAM50A | 9130 | 203262_s_at | 1.0039 | 0.0017 | |
| GSE97050 | FAM50A | 9130 | A_23_P34107 | 0.1422 | 0.6212 | |
| SRP007169 | FAM50A | 9130 | RNAseq | 0.9666 | 0.1206 | |
| SRP064894 | FAM50A | 9130 | RNAseq | 0.9263 | 0.0005 | |
| SRP133303 | FAM50A | 9130 | RNAseq | 0.8899 | 0.0001 | |
| SRP159526 | FAM50A | 9130 | RNAseq | 0.6444 | 0.0124 | |
| SRP193095 | FAM50A | 9130 | RNAseq | 0.5272 | 0.0001 | |
| SRP219564 | FAM50A | 9130 | RNAseq | 0.6561 | 0.1340 | |
| TCGA | FAM50A | 9130 | RNAseq | 0.0034 | 0.9545 |
Upregulated datasets: 1; Downregulated datasets: 0.
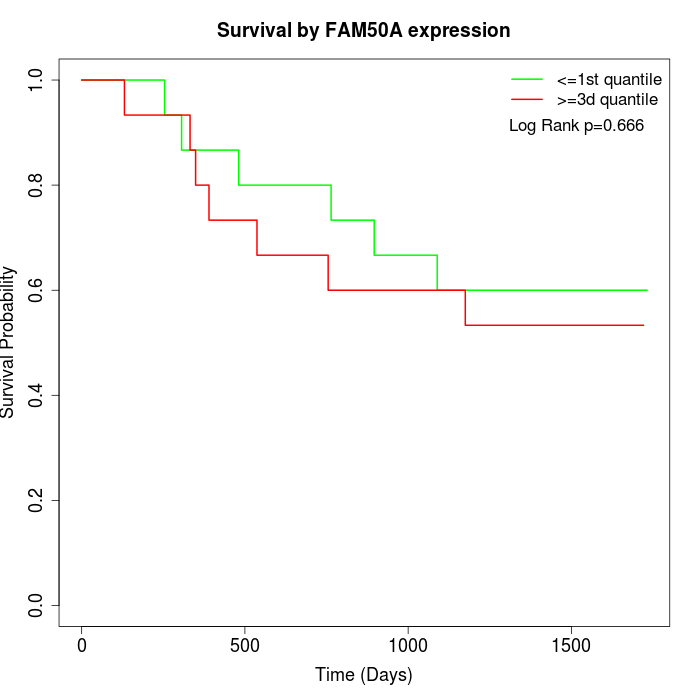
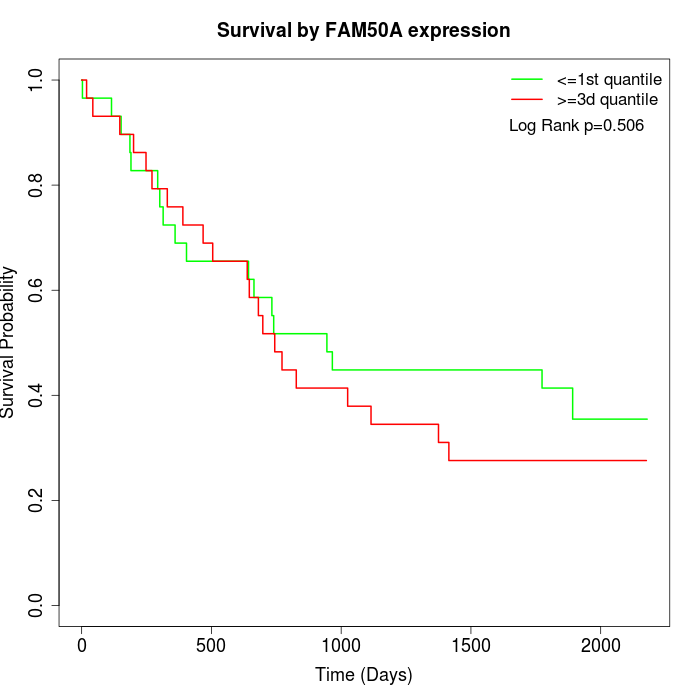
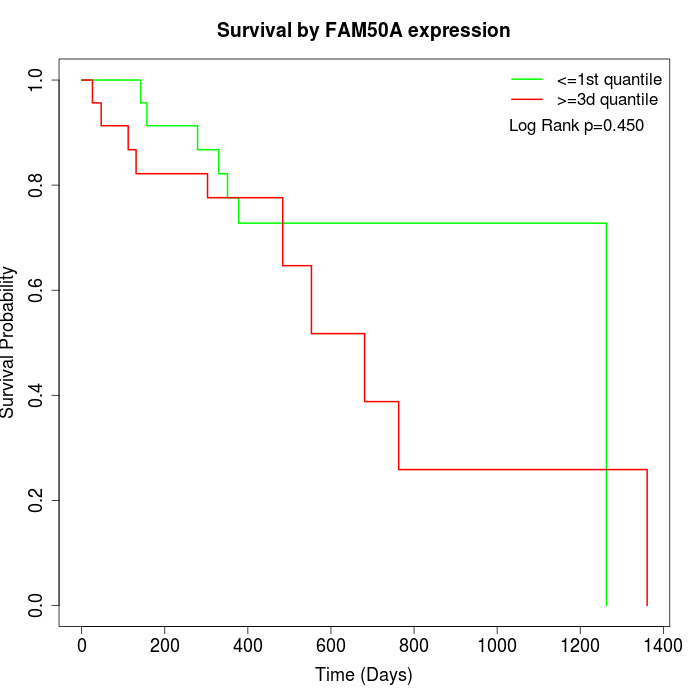
Survival by FAM50A expression:
Note: Click image to view full size file.
Copy number change of FAM50A:
No record found for this gene.
Somatic mutations of FAM50A:
Generating mutation plots.
Highly correlated genes for FAM50A:
Showing top 20/763 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FAM50A | ALKBH6 | 0.798391 | 3 | 0 | 3 |
| FAM50A | PCDH18 | 0.773832 | 3 | 0 | 3 |
| FAM50A | SESTD1 | 0.76734 | 3 | 0 | 3 |
| FAM50A | WDR81 | 0.751763 | 4 | 0 | 4 |
| FAM50A | TAF1C | 0.744372 | 3 | 0 | 3 |
| FAM50A | STAT5B | 0.738247 | 4 | 0 | 4 |
| FAM50A | INTS3 | 0.730441 | 4 | 0 | 3 |
| FAM50A | SDHAF2 | 0.726876 | 3 | 0 | 3 |
| FAM50A | MSL1 | 0.726532 | 3 | 0 | 3 |
| FAM50A | SPIRE1 | 0.724907 | 4 | 0 | 4 |
| FAM50A | NR2F2 | 0.71871 | 3 | 0 | 3 |
| FAM50A | SELE | 0.717066 | 3 | 0 | 3 |
| FAM50A | CDC26 | 0.708736 | 3 | 0 | 3 |
| FAM50A | CAMKK1 | 0.707483 | 3 | 0 | 3 |
| FAM50A | SSBP4 | 0.705584 | 4 | 0 | 3 |
| FAM50A | NCOR2 | 0.702014 | 7 | 0 | 7 |
| FAM50A | MRPS26 | 0.701916 | 4 | 0 | 4 |
| FAM50A | SLC26A11 | 0.699992 | 4 | 0 | 4 |
| FAM50A | HERC2 | 0.697593 | 3 | 0 | 3 |
| FAM50A | LSM14B | 0.695736 | 3 | 0 | 3 |
For details and further investigation, click here