| Full name: neuronal pentraxin 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 7q22.1 | ||
| Entrez ID: 4885 | HGNC ID: HGNC:7953 | Ensembl Gene: ENSG00000106236 | OMIM ID: 600750 |
| Drug and gene relationship at DGIdb | |||
Expression of NPTX2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NPTX2 | 4885 | 213479_at | -0.5239 | 0.7122 | |
| GSE20347 | NPTX2 | 4885 | 213479_at | 0.7334 | 0.1710 | |
| GSE23400 | NPTX2 | 4885 | 213479_at | 0.0988 | 0.2186 | |
| GSE26886 | NPTX2 | 4885 | 213479_at | 1.3238 | 0.0033 | |
| GSE29001 | NPTX2 | 4885 | 213479_at | 1.0913 | 0.2020 | |
| GSE38129 | NPTX2 | 4885 | 213479_at | 0.3008 | 0.5518 | |
| GSE45670 | NPTX2 | 4885 | 213479_at | -0.1916 | 0.5953 | |
| GSE53622 | NPTX2 | 4885 | 22453 | 0.7545 | 0.0289 | |
| GSE53624 | NPTX2 | 4885 | 22453 | 0.7862 | 0.0005 | |
| GSE63941 | NPTX2 | 4885 | 213479_at | 0.1442 | 0.9081 | |
| GSE77861 | NPTX2 | 4885 | 213479_at | 0.6952 | 0.1911 | |
| SRP064894 | NPTX2 | 4885 | RNAseq | 1.6395 | 0.0003 | |
| SRP219564 | NPTX2 | 4885 | RNAseq | 0.1216 | 0.8802 | |
| TCGA | NPTX2 | 4885 | RNAseq | -0.4159 | 0.1592 |
Upregulated datasets: 2; Downregulated datasets: 0.
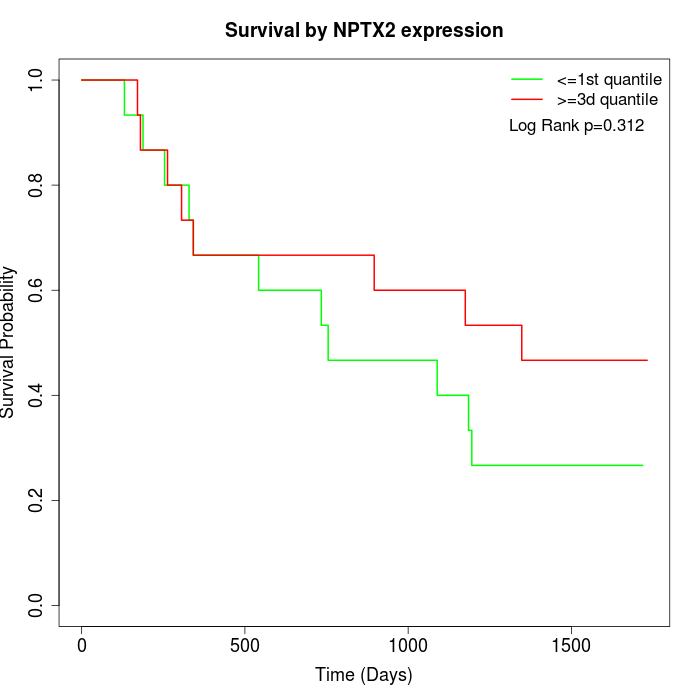
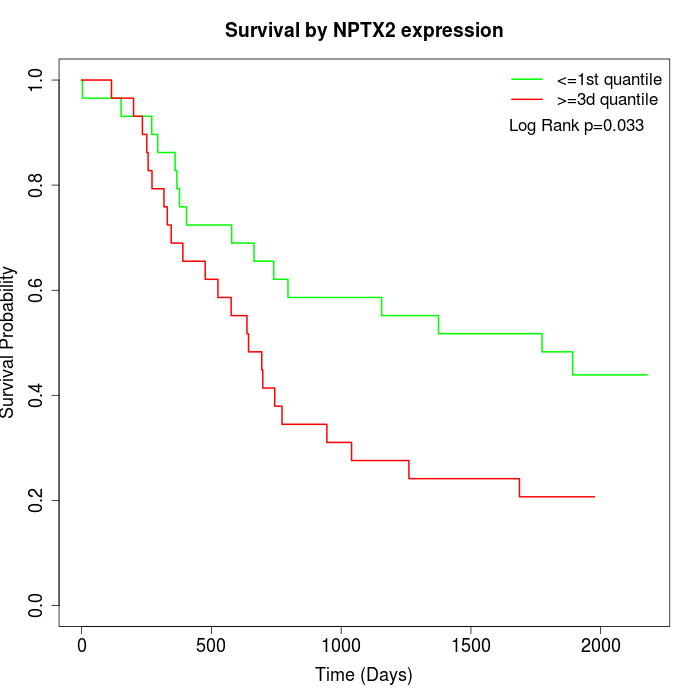
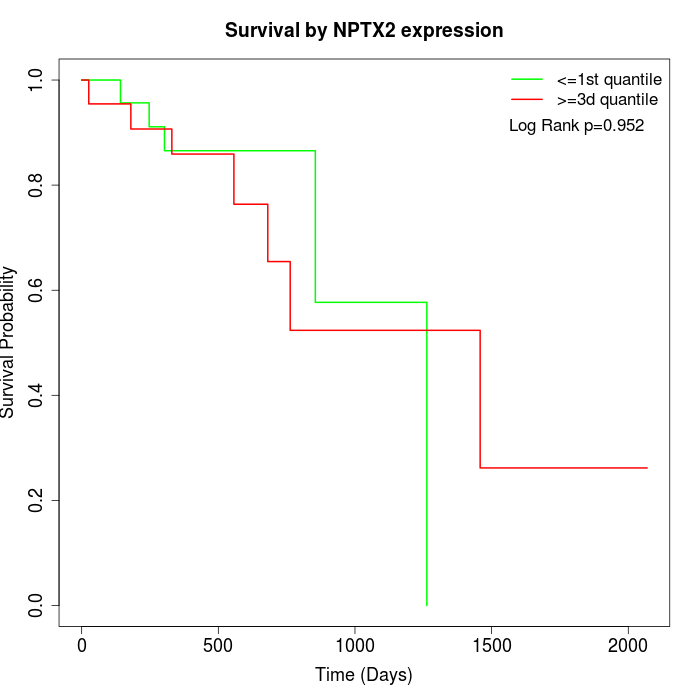
Survival by NPTX2 expression:
Note: Click image to view full size file.
Copy number change of NPTX2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NPTX2 | 4885 | 13 | 0 | 17 | |
| GSE20123 | NPTX2 | 4885 | 12 | 0 | 18 | |
| GSE43470 | NPTX2 | 4885 | 6 | 1 | 36 | |
| GSE46452 | NPTX2 | 4885 | 12 | 0 | 47 | |
| GSE47630 | NPTX2 | 4885 | 8 | 3 | 29 | |
| GSE54993 | NPTX2 | 4885 | 1 | 9 | 60 | |
| GSE54994 | NPTX2 | 4885 | 15 | 3 | 35 | |
| GSE60625 | NPTX2 | 4885 | 0 | 0 | 11 | |
| GSE74703 | NPTX2 | 4885 | 6 | 1 | 29 | |
| GSE74704 | NPTX2 | 4885 | 9 | 0 | 11 | |
| TCGA | NPTX2 | 4885 | 54 | 4 | 38 |
Total number of gains: 136; Total number of losses: 21; Total Number of normals: 331.
Somatic mutations of NPTX2:
Generating mutation plots.
Highly correlated genes for NPTX2:
Showing top 20/313 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NPTX2 | FGF3 | 0.799217 | 6 | 0 | 6 |
| NPTX2 | MYBPC1 | 0.787934 | 4 | 0 | 4 |
| NPTX2 | ARNT2 | 0.773104 | 6 | 0 | 6 |
| NPTX2 | FILIP1 | 0.770799 | 3 | 0 | 3 |
| NPTX2 | CCDC8 | 0.769792 | 3 | 0 | 3 |
| NPTX2 | ADD2 | 0.760191 | 3 | 0 | 3 |
| NPTX2 | EPHA7 | 0.697096 | 4 | 0 | 3 |
| NPTX2 | LIX1 | 0.694639 | 3 | 0 | 3 |
| NPTX2 | PPP1R12A | 0.686897 | 3 | 0 | 3 |
| NPTX2 | BEX1 | 0.686517 | 6 | 0 | 4 |
| NPTX2 | NGFR | 0.679594 | 6 | 0 | 4 |
| NPTX2 | NRXN3 | 0.671317 | 3 | 0 | 3 |
| NPTX2 | ICAM2 | 0.667976 | 4 | 0 | 3 |
| NPTX2 | CSN3 | 0.657394 | 4 | 0 | 3 |
| NPTX2 | CRABP1 | 0.654446 | 5 | 0 | 5 |
| NPTX2 | GFRA1 | 0.653067 | 4 | 0 | 3 |
| NPTX2 | ATP8A2 | 0.652722 | 7 | 0 | 6 |
| NPTX2 | DLC1 | 0.651631 | 4 | 0 | 3 |
| NPTX2 | COL4A6 | 0.650439 | 5 | 0 | 4 |
| NPTX2 | NRXN1 | 0.648193 | 5 | 0 | 3 |
For details and further investigation, click here