| Full name: galectin 4 | Alias Symbol: GAL4 | ||
| Type: protein-coding gene | Cytoband: 19q13.2 | ||
| Entrez ID: 3960 | HGNC ID: HGNC:6565 | Ensembl Gene: ENSG00000171747 | OMIM ID: 602518 |
| Drug and gene relationship at DGIdb | |||
Expression of LGALS4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LGALS4 | 3960 | 204272_at | -0.0378 | 0.9228 | |
| GSE20347 | LGALS4 | 3960 | 204272_at | -0.0272 | 0.7966 | |
| GSE23400 | LGALS4 | 3960 | 204272_at | -0.2243 | 0.0362 | |
| GSE26886 | LGALS4 | 3960 | 204272_at | 0.2742 | 0.3268 | |
| GSE29001 | LGALS4 | 3960 | 204272_at | 0.1086 | 0.8107 | |
| GSE38129 | LGALS4 | 3960 | 204272_at | -0.4824 | 0.0792 | |
| GSE45670 | LGALS4 | 3960 | 204272_at | 0.0299 | 0.8206 | |
| GSE53622 | LGALS4 | 3960 | 37836 | -0.0374 | 0.9196 | |
| GSE53624 | LGALS4 | 3960 | 37836 | -0.9417 | 0.0000 | |
| GSE63941 | LGALS4 | 3960 | 204272_at | 0.1514 | 0.3971 | |
| GSE77861 | LGALS4 | 3960 | 204272_at | -0.0340 | 0.7965 | |
| SRP219564 | LGALS4 | 3960 | RNAseq | -1.2813 | 0.0062 | |
| TCGA | LGALS4 | 3960 | RNAseq | -2.9156 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 2.
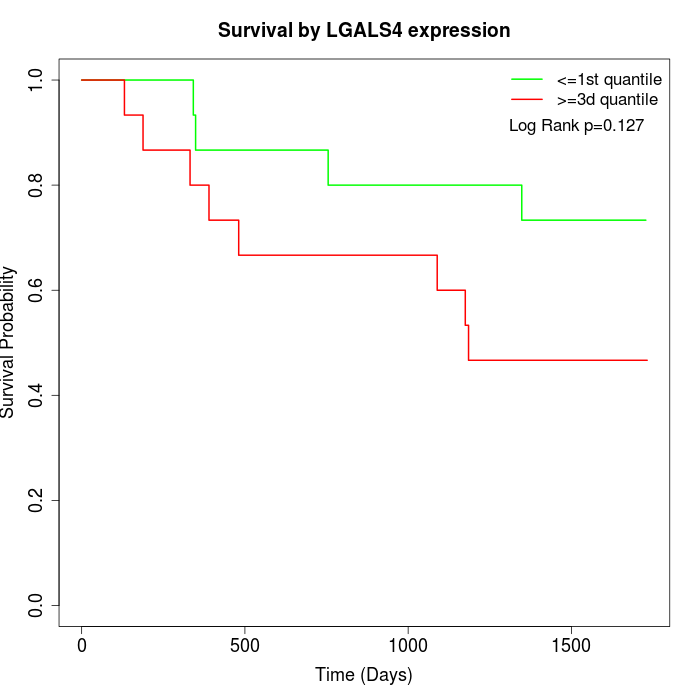
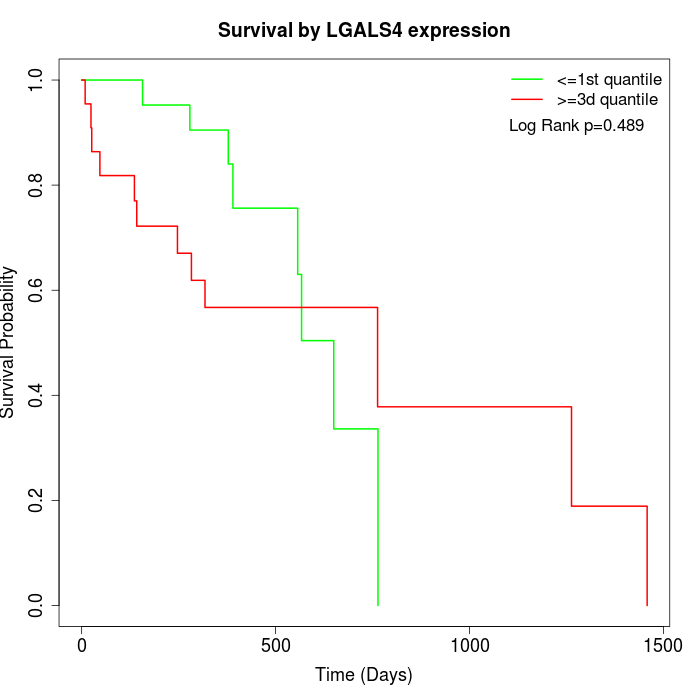
Survival by LGALS4 expression:
Note: Click image to view full size file.
Copy number change of LGALS4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LGALS4 | 3960 | 7 | 4 | 19 | |
| GSE20123 | LGALS4 | 3960 | 7 | 3 | 20 | |
| GSE43470 | LGALS4 | 3960 | 4 | 10 | 29 | |
| GSE46452 | LGALS4 | 3960 | 47 | 2 | 10 | |
| GSE47630 | LGALS4 | 3960 | 9 | 5 | 26 | |
| GSE54993 | LGALS4 | 3960 | 17 | 3 | 50 | |
| GSE54994 | LGALS4 | 3960 | 6 | 9 | 38 | |
| GSE60625 | LGALS4 | 3960 | 9 | 0 | 2 | |
| GSE74703 | LGALS4 | 3960 | 4 | 6 | 26 | |
| GSE74704 | LGALS4 | 3960 | 7 | 1 | 12 | |
| TCGA | LGALS4 | 3960 | 15 | 16 | 65 |
Total number of gains: 132; Total number of losses: 59; Total Number of normals: 297.
Somatic mutations of LGALS4:
Generating mutation plots.
Highly correlated genes for LGALS4:
Showing top 20/84 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LGALS4 | CTSE | 0.709111 | 6 | 0 | 5 |
| LGALS4 | SPINK1 | 0.678619 | 5 | 0 | 4 |
| LGALS4 | KCNE2 | 0.670474 | 5 | 0 | 4 |
| LGALS4 | LIPF | 0.6649 | 5 | 0 | 4 |
| LGALS4 | KRT20 | 0.66427 | 5 | 0 | 4 |
| LGALS4 | GKN2 | 0.651989 | 4 | 0 | 3 |
| LGALS4 | TPH1 | 0.651131 | 3 | 0 | 3 |
| LGALS4 | FUT9 | 0.64926 | 5 | 0 | 4 |
| LGALS4 | PLA2G1B | 0.640861 | 4 | 0 | 3 |
| LGALS4 | DRD2 | 0.640786 | 3 | 0 | 3 |
| LGALS4 | GKN1 | 0.639781 | 7 | 0 | 6 |
| LGALS4 | SERPINA4 | 0.633567 | 6 | 0 | 5 |
| LGALS4 | TFF2 | 0.630081 | 6 | 0 | 4 |
| LGALS4 | ATP4A | 0.628762 | 6 | 0 | 4 |
| LGALS4 | PHGR1 | 0.625849 | 4 | 0 | 3 |
| LGALS4 | GATA6-AS1 | 0.624571 | 4 | 0 | 3 |
| LGALS4 | PGC | 0.624154 | 6 | 0 | 4 |
| LGALS4 | ATP4B | 0.621811 | 6 | 0 | 4 |
| LGALS4 | CPA2 | 0.616059 | 6 | 0 | 4 |
| LGALS4 | UGT2B15 | 0.612302 | 6 | 0 | 6 |
For details and further investigation, click here