| Full name: galactosidase alpha | Alias Symbol: GALA | ||
| Type: protein-coding gene | Cytoband: Xq22.1 | ||
| Entrez ID: 2717 | HGNC ID: HGNC:4296 | Ensembl Gene: ENSG00000102393 | OMIM ID: 300644 |
| Drug and gene relationship at DGIdb | |||
Expression of GLA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GLA | 2717 | 214430_at | 0.5868 | 0.1359 | |
| GSE20347 | GLA | 2717 | 214430_at | 0.9494 | 0.0000 | |
| GSE23400 | GLA | 2717 | 214430_at | 0.6082 | 0.0000 | |
| GSE26886 | GLA | 2717 | 214430_at | 0.8716 | 0.0001 | |
| GSE29001 | GLA | 2717 | 214430_at | 0.6170 | 0.0733 | |
| GSE38129 | GLA | 2717 | 214430_at | 0.9730 | 0.0000 | |
| GSE45670 | GLA | 2717 | 214430_at | 0.8352 | 0.0035 | |
| GSE53622 | GLA | 2717 | 47204 | 0.9190 | 0.0000 | |
| GSE53624 | GLA | 2717 | 47204 | 1.1429 | 0.0000 | |
| GSE63941 | GLA | 2717 | 214430_at | -0.4253 | 0.4631 | |
| GSE77861 | GLA | 2717 | 214430_at | 1.2763 | 0.0035 | |
| GSE97050 | GLA | 2717 | A_23_P45475 | 0.2202 | 0.5121 | |
| SRP007169 | GLA | 2717 | RNAseq | 1.1838 | 0.0110 | |
| SRP008496 | GLA | 2717 | RNAseq | 1.3760 | 0.0000 | |
| SRP064894 | GLA | 2717 | RNAseq | 1.0134 | 0.0000 | |
| SRP133303 | GLA | 2717 | RNAseq | 1.1089 | 0.0000 | |
| SRP159526 | GLA | 2717 | RNAseq | 1.2627 | 0.0000 | |
| SRP193095 | GLA | 2717 | RNAseq | 0.8961 | 0.0000 | |
| SRP219564 | GLA | 2717 | RNAseq | 0.8410 | 0.1695 | |
| TCGA | GLA | 2717 | RNAseq | 0.4026 | 0.0000 |
Upregulated datasets: 7; Downregulated datasets: 0.
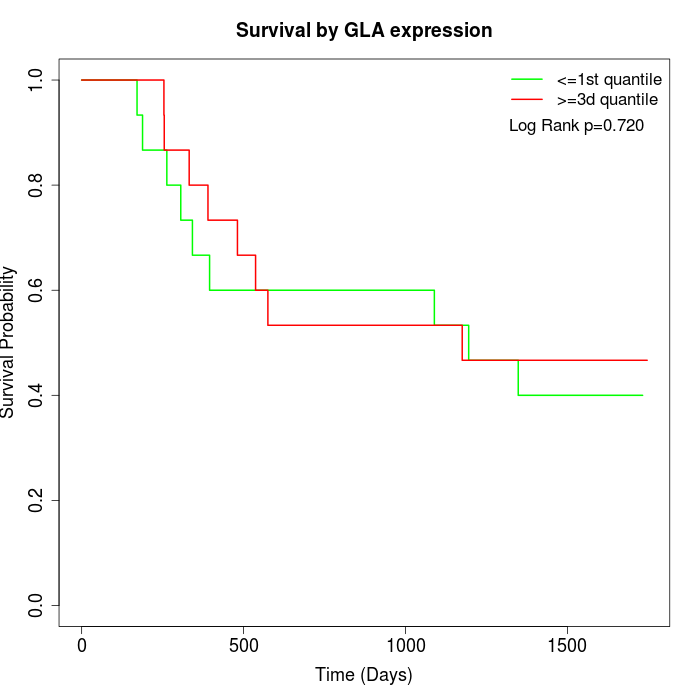
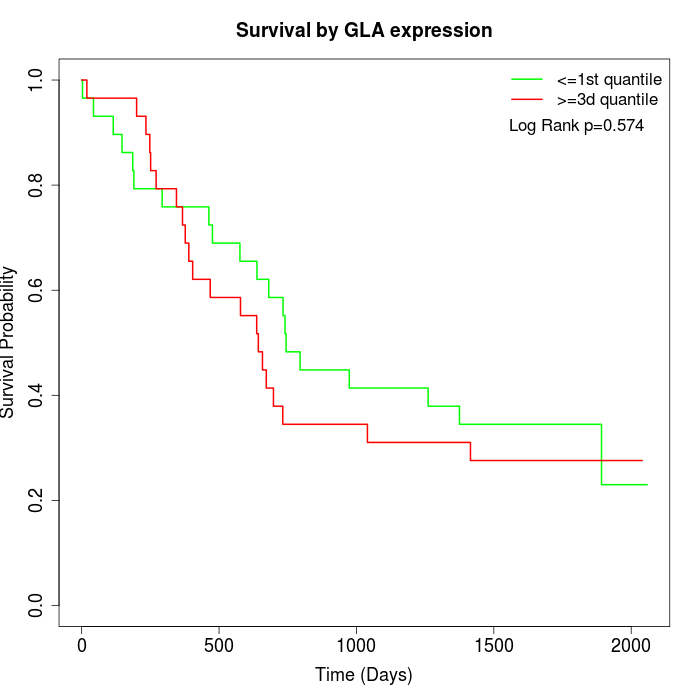
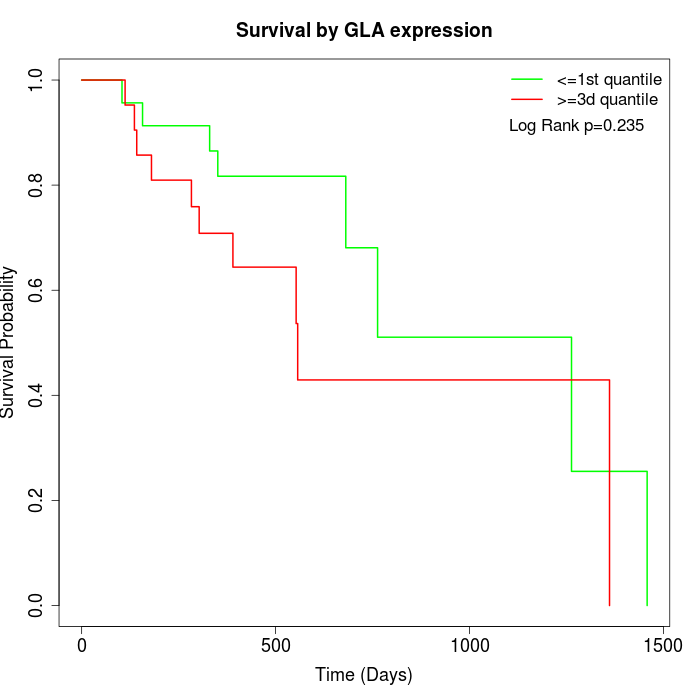
Survival by GLA expression:
Note: Click image to view full size file.
Copy number change of GLA:
No record found for this gene.
Somatic mutations of GLA:
Generating mutation plots.
Highly correlated genes for GLA:
Showing top 20/1733 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GLA | DOCK11 | 0.790845 | 3 | 0 | 3 |
| GLA | SNTB2 | 0.771607 | 3 | 0 | 3 |
| GLA | TBC1D8B | 0.767398 | 3 | 0 | 3 |
| GLA | TTC39C | 0.761654 | 3 | 0 | 3 |
| GLA | ZCCHC7 | 0.758514 | 3 | 0 | 3 |
| GLA | SPRR2G | 0.752885 | 3 | 0 | 3 |
| GLA | COA6 | 0.752695 | 6 | 0 | 6 |
| GLA | FAM89A | 0.733746 | 3 | 0 | 3 |
| GLA | HAS3 | 0.731122 | 5 | 0 | 4 |
| GLA | SOCS4 | 0.722981 | 4 | 0 | 4 |
| GLA | SGTB | 0.719065 | 4 | 0 | 3 |
| GLA | ASF1A | 0.716521 | 4 | 0 | 3 |
| GLA | WDR66 | 0.714617 | 6 | 0 | 6 |
| GLA | FAM89B | 0.710609 | 3 | 0 | 3 |
| GLA | PTGS2 | 0.710314 | 3 | 0 | 3 |
| GLA | CDCA2 | 0.705606 | 5 | 0 | 5 |
| GLA | DDX39A | 0.704554 | 11 | 0 | 11 |
| GLA | FIP1L1 | 0.703452 | 3 | 0 | 3 |
| GLA | CCDC86 | 0.701105 | 8 | 0 | 8 |
| GLA | NEIL2 | 0.700451 | 5 | 0 | 5 |
For details and further investigation, click here