| Full name: glycine-N-acyltransferase like 1 | Alias Symbol: MGC15397|FLJ34646 | ||
| Type: protein-coding gene | Cytoband: 11q12.1 | ||
| Entrez ID: 92292 | HGNC ID: HGNC:30519 | Ensembl Gene: ENSG00000166840 | OMIM ID: 614761 |
| Drug and gene relationship at DGIdb | |||
Expression of GLYATL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GLYATL1 | 92292 | 1562089_at | -0.0656 | 0.7350 | |
| GSE26886 | GLYATL1 | 92292 | 1562089_at | -0.0097 | 0.9148 | |
| GSE45670 | GLYATL1 | 92292 | 1562089_at | -0.0631 | 0.3563 | |
| GSE53622 | GLYATL1 | 92292 | 19279 | -0.1646 | 0.4930 | |
| GSE53624 | GLYATL1 | 92292 | 19279 | -0.7095 | 0.0000 | |
| GSE63941 | GLYATL1 | 92292 | 1562089_at | 0.1274 | 0.3292 | |
| GSE77861 | GLYATL1 | 92292 | 1562089_at | -0.0477 | 0.5704 | |
| GSE97050 | GLYATL1 | A_21_P0010977 | 0.1443 | 0.5335 | ||
| SRP133303 | GLYATL1 | 92292 | RNAseq | -0.2795 | 0.4038 | |
| TCGA | GLYATL1 | 92292 | RNAseq | -3.3394 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
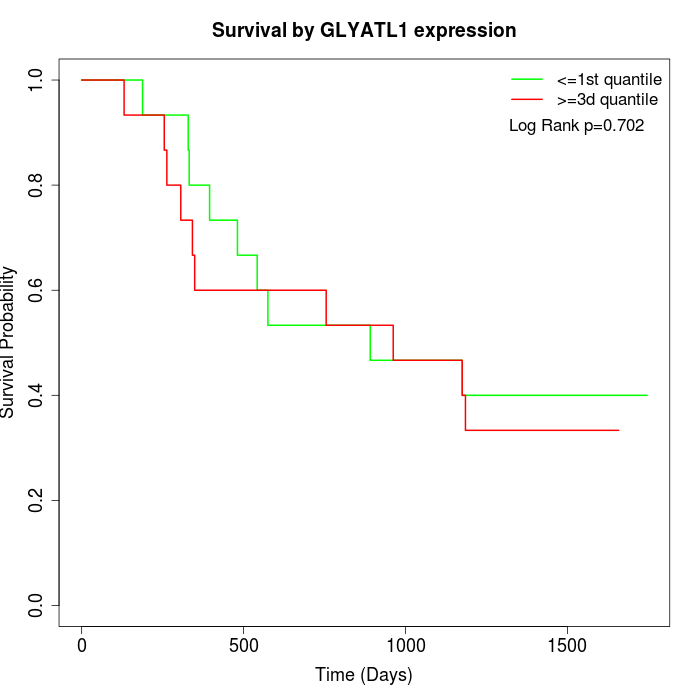
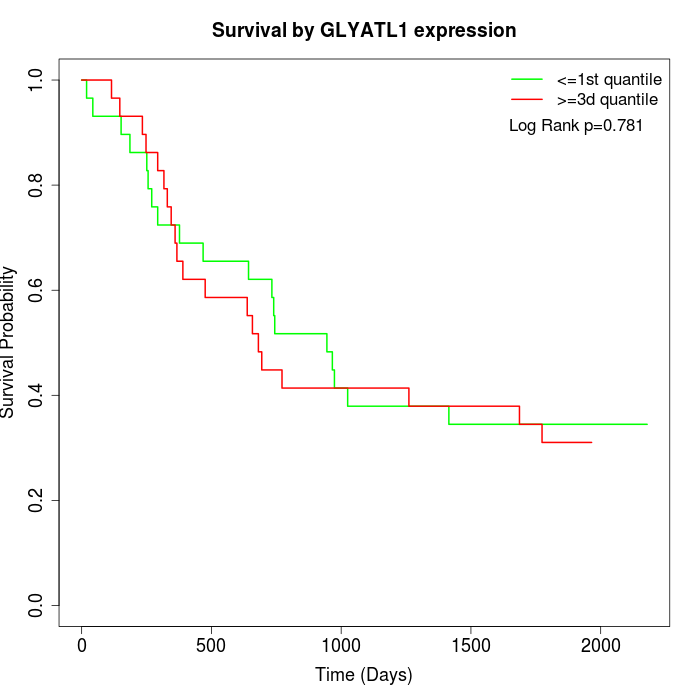
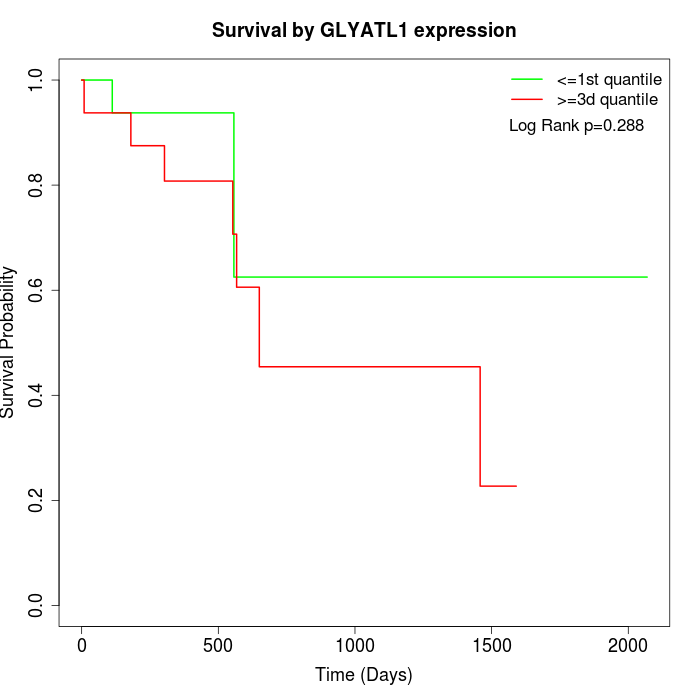
Survival by GLYATL1 expression:
Note: Click image to view full size file.
Copy number change of GLYATL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GLYATL1 | 92292 | 1 | 5 | 24 | |
| GSE20123 | GLYATL1 | 92292 | 1 | 5 | 24 | |
| GSE43470 | GLYATL1 | 92292 | 1 | 5 | 37 | |
| GSE46452 | GLYATL1 | 92292 | 10 | 4 | 45 | |
| GSE47630 | GLYATL1 | 92292 | 3 | 8 | 29 | |
| GSE54993 | GLYATL1 | 92292 | 3 | 0 | 67 | |
| GSE54994 | GLYATL1 | 92292 | 5 | 6 | 42 | |
| GSE60625 | GLYATL1 | 92292 | 0 | 3 | 8 | |
| GSE74703 | GLYATL1 | 92292 | 1 | 3 | 32 | |
| GSE74704 | GLYATL1 | 92292 | 1 | 4 | 15 | |
| TCGA | GLYATL1 | 92292 | 14 | 10 | 72 |
Total number of gains: 40; Total number of losses: 53; Total Number of normals: 395.
Somatic mutations of GLYATL1:
Generating mutation plots.
Highly correlated genes for GLYATL1:
Showing top 20/34 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GLYATL1 | TRIM43 | 0.756151 | 3 | 0 | 3 |
| GLYATL1 | SLC5A4 | 0.741933 | 3 | 0 | 3 |
| GLYATL1 | MEP1A | 0.729517 | 3 | 0 | 3 |
| GLYATL1 | LY6G6C | 0.717075 | 3 | 0 | 3 |
| GLYATL1 | HEATR4 | 0.686479 | 3 | 0 | 3 |
| GLYATL1 | GPAT2 | 0.685199 | 3 | 0 | 3 |
| GLYATL1 | RP1L1 | 0.672842 | 3 | 0 | 3 |
| GLYATL1 | ABCD3 | 0.671694 | 4 | 0 | 3 |
| GLYATL1 | FOXA3 | 0.669747 | 3 | 0 | 3 |
| GLYATL1 | PRG3 | 0.665366 | 3 | 0 | 3 |
| GLYATL1 | BCL2L10 | 0.66506 | 3 | 0 | 3 |
| GLYATL1 | HPX | 0.657045 | 3 | 0 | 3 |
| GLYATL1 | MMP21 | 0.656839 | 3 | 0 | 3 |
| GLYATL1 | SH2D1B | 0.640261 | 4 | 0 | 3 |
| GLYATL1 | MYOZ3 | 0.637678 | 3 | 0 | 3 |
| GLYATL1 | WDR87 | 0.631485 | 4 | 0 | 4 |
| GLYATL1 | GRM1 | 0.630123 | 3 | 0 | 3 |
| GLYATL1 | SPTA1 | 0.61525 | 4 | 0 | 3 |
| GLYATL1 | CD300LG | 0.614806 | 4 | 0 | 3 |
| GLYATL1 | PRSS2 | 0.600347 | 3 | 0 | 3 |
For details and further investigation, click here