| Full name: GNAS antisense RNA 1 | Alias Symbol: SANG|NESP-AS|NESPAS|GNAS1AS|NCRNA00075 | ||
| Type: non-coding RNA | Cytoband: 20q13.32 | ||
| Entrez ID: 149775 | HGNC ID: HGNC:24872 | Ensembl Gene: ENSG00000235590 | OMIM ID: 610540 |
| Drug and gene relationship at DGIdb | |||
Expression of GNAS-AS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GNAS-AS1 | 149775 | 232881_at | 0.6320 | 0.4558 | |
| GSE26886 | GNAS-AS1 | 149775 | 232881_at | 0.2752 | 0.0720 | |
| GSE45670 | GNAS-AS1 | 149775 | 232881_at | 0.1501 | 0.4520 | |
| GSE53622 | GNAS-AS1 | 149775 | 42275 | 0.6973 | 0.0000 | |
| GSE53624 | GNAS-AS1 | 149775 | 42275 | 0.4119 | 0.0012 | |
| GSE63941 | GNAS-AS1 | 149775 | 232881_at | -0.3635 | 0.1816 | |
| GSE77861 | GNAS-AS1 | 149775 | 232881_at | -0.1140 | 0.2371 | |
| SRP133303 | GNAS-AS1 | 149775 | RNAseq | -0.9584 | 0.0374 |
Upregulated datasets: 0; Downregulated datasets: 0.
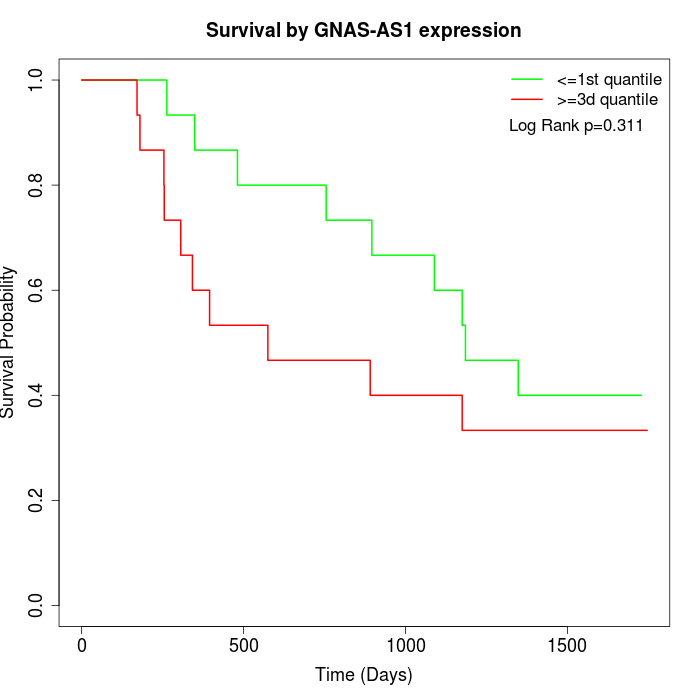
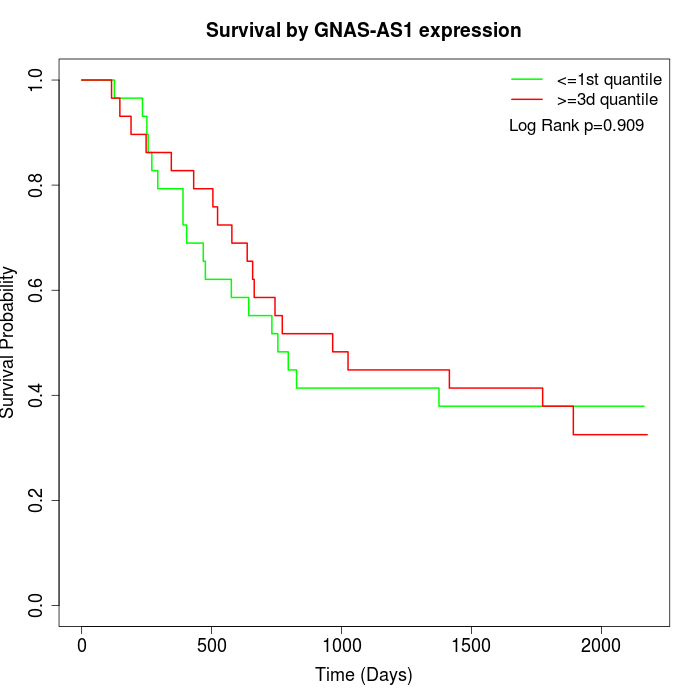
Survival by GNAS-AS1 expression:
Note: Click image to view full size file.
Copy number change of GNAS-AS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GNAS-AS1 | 149775 | 14 | 2 | 14 | |
| GSE20123 | GNAS-AS1 | 149775 | 14 | 2 | 14 | |
| GSE43470 | GNAS-AS1 | 149775 | 13 | 0 | 30 | |
| GSE46452 | GNAS-AS1 | 149775 | 29 | 0 | 30 | |
| GSE47630 | GNAS-AS1 | 149775 | 24 | 1 | 15 | |
| GSE54993 | GNAS-AS1 | 149775 | 0 | 17 | 53 | |
| GSE54994 | GNAS-AS1 | 149775 | 29 | 0 | 24 | |
| GSE60625 | GNAS-AS1 | 149775 | 0 | 0 | 11 | |
| GSE74703 | GNAS-AS1 | 149775 | 11 | 0 | 25 | |
| GSE74704 | GNAS-AS1 | 149775 | 11 | 0 | 9 |
Total number of gains: 145; Total number of losses: 22; Total Number of normals: 225.
Somatic mutations of GNAS-AS1:
Generating mutation plots.
Highly correlated genes for GNAS-AS1:
Showing top 20/32 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GNAS-AS1 | AP4S1 | 0.740401 | 3 | 0 | 3 |
| GNAS-AS1 | DUSP15 | 0.663539 | 3 | 0 | 3 |
| GNAS-AS1 | HOXD9 | 0.659564 | 3 | 0 | 3 |
| GNAS-AS1 | CLMN | 0.656084 | 3 | 0 | 3 |
| GNAS-AS1 | HOXD11 | 0.648318 | 3 | 0 | 3 |
| GNAS-AS1 | TG | 0.64801 | 3 | 0 | 3 |
| GNAS-AS1 | SMIM2 | 0.646566 | 3 | 0 | 3 |
| GNAS-AS1 | HOXD10 | 0.641897 | 3 | 0 | 3 |
| GNAS-AS1 | TCOF1 | 0.621351 | 3 | 0 | 3 |
| GNAS-AS1 | TMEM91 | 0.608052 | 3 | 0 | 3 |
| GNAS-AS1 | LRRC56 | 0.589002 | 3 | 0 | 3 |
| GNAS-AS1 | CEP128 | 0.588929 | 4 | 0 | 3 |
| GNAS-AS1 | GTSE1 | 0.588364 | 4 | 0 | 3 |
| GNAS-AS1 | MYO7A | 0.585844 | 3 | 0 | 3 |
| GNAS-AS1 | CHAF1A | 0.583904 | 4 | 0 | 3 |
| GNAS-AS1 | PLEKHA4 | 0.579638 | 3 | 0 | 3 |
| GNAS-AS1 | FANCA | 0.577273 | 4 | 0 | 3 |
| GNAS-AS1 | DLX4 | 0.576977 | 3 | 0 | 3 |
| GNAS-AS1 | CSNK1G2 | 0.562345 | 3 | 0 | 3 |
| GNAS-AS1 | SHPK | 0.557268 | 3 | 0 | 3 |
For details and further investigation, click here