| Full name: EPH receptor B4 | Alias Symbol: Tyro11 | ||
| Type: protein-coding gene | Cytoband: 7q22.1 | ||
| Entrez ID: 2050 | HGNC ID: HGNC:3395 | Ensembl Gene: ENSG00000196411 | OMIM ID: 600011 |
| Drug and gene relationship at DGIdb | |||
EPHB4 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance |
Expression of EPHB4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EPHB4 | 2050 | 202894_at | 0.8750 | 0.0143 | |
| GSE20347 | EPHB4 | 2050 | 202894_at | 0.9710 | 0.0000 | |
| GSE23400 | EPHB4 | 2050 | 202894_at | 0.7400 | 0.0000 | |
| GSE26886 | EPHB4 | 2050 | 202894_at | 1.5858 | 0.0000 | |
| GSE29001 | EPHB4 | 2050 | 202894_at | 1.0676 | 0.0079 | |
| GSE38129 | EPHB4 | 2050 | 202894_at | 0.8234 | 0.0000 | |
| GSE45670 | EPHB4 | 2050 | 202894_at | 0.5743 | 0.0046 | |
| GSE53622 | EPHB4 | 2050 | 109182 | 1.0088 | 0.0000 | |
| GSE53624 | EPHB4 | 2050 | 109182 | 0.6620 | 0.0000 | |
| GSE63941 | EPHB4 | 2050 | 202894_at | -0.1934 | 0.6451 | |
| GSE77861 | EPHB4 | 2050 | 202894_at | 0.6860 | 0.0126 | |
| GSE97050 | EPHB4 | 2050 | A_33_P3216532 | 0.5781 | 0.1983 | |
| SRP007169 | EPHB4 | 2050 | RNAseq | 1.0580 | 0.0030 | |
| SRP008496 | EPHB4 | 2050 | RNAseq | 1.3626 | 0.0000 | |
| SRP064894 | EPHB4 | 2050 | RNAseq | 1.0517 | 0.0000 | |
| SRP133303 | EPHB4 | 2050 | RNAseq | 0.7638 | 0.0070 | |
| SRP159526 | EPHB4 | 2050 | RNAseq | 1.0882 | 0.0042 | |
| SRP193095 | EPHB4 | 2050 | RNAseq | 1.2604 | 0.0000 | |
| SRP219564 | EPHB4 | 2050 | RNAseq | 0.8997 | 0.0332 | |
| TCGA | EPHB4 | 2050 | RNAseq | 0.1955 | 0.0064 |
Upregulated datasets: 8; Downregulated datasets: 0.
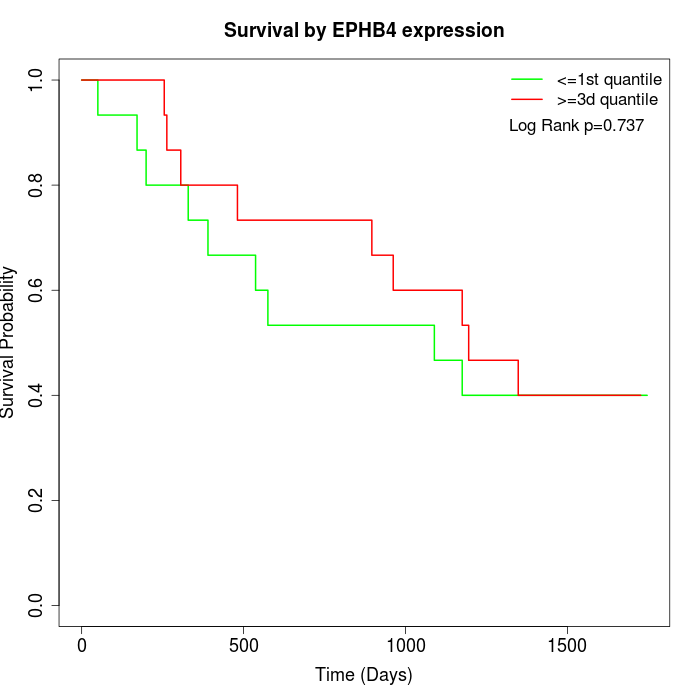
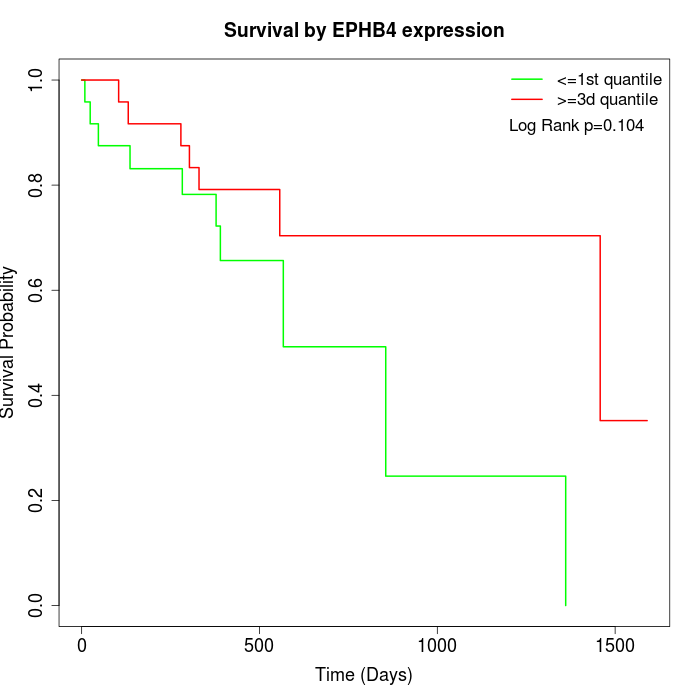
Survival by EPHB4 expression:
Note: Click image to view full size file.
Copy number change of EPHB4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EPHB4 | 2050 | 13 | 0 | 17 | |
| GSE20123 | EPHB4 | 2050 | 13 | 0 | 17 | |
| GSE43470 | EPHB4 | 2050 | 7 | 2 | 34 | |
| GSE46452 | EPHB4 | 2050 | 11 | 1 | 47 | |
| GSE47630 | EPHB4 | 2050 | 7 | 3 | 30 | |
| GSE54993 | EPHB4 | 2050 | 1 | 9 | 60 | |
| GSE54994 | EPHB4 | 2050 | 16 | 3 | 34 | |
| GSE60625 | EPHB4 | 2050 | 0 | 0 | 11 | |
| GSE74703 | EPHB4 | 2050 | 7 | 1 | 28 | |
| GSE74704 | EPHB4 | 2050 | 9 | 0 | 11 | |
| TCGA | EPHB4 | 2050 | 53 | 6 | 37 |
Total number of gains: 137; Total number of losses: 25; Total Number of normals: 326.
Somatic mutations of EPHB4:
Generating mutation plots.
Highly correlated genes for EPHB4:
Showing top 20/1904 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EPHB4 | EME1 | 0.782465 | 5 | 0 | 5 |
| EPHB4 | POU5F2 | 0.775353 | 3 | 0 | 3 |
| EPHB4 | MYBL2 | 0.755883 | 11 | 0 | 11 |
| EPHB4 | HCFC1R1 | 0.755813 | 4 | 0 | 4 |
| EPHB4 | TMEM132A | 0.734291 | 9 | 0 | 9 |
| EPHB4 | CDCA2 | 0.733483 | 5 | 0 | 5 |
| EPHB4 | MCM6 | 0.731293 | 9 | 0 | 9 |
| EPHB4 | MYO10 | 0.729059 | 7 | 0 | 7 |
| EPHB4 | UBE2T | 0.725903 | 7 | 0 | 6 |
| EPHB4 | SCYL1 | 0.722699 | 4 | 0 | 4 |
| EPHB4 | PPP1R35 | 0.720805 | 7 | 0 | 6 |
| EPHB4 | APMAP | 0.718389 | 9 | 0 | 9 |
| EPHB4 | UBE2C | 0.713652 | 11 | 0 | 11 |
| EPHB4 | CDC25B | 0.713381 | 11 | 0 | 10 |
| EPHB4 | PLOD3 | 0.710685 | 12 | 0 | 10 |
| EPHB4 | DNMT1 | 0.710171 | 11 | 0 | 10 |
| EPHB4 | ARPC1B | 0.710126 | 10 | 0 | 9 |
| EPHB4 | MRPL14 | 0.709396 | 4 | 0 | 3 |
| EPHB4 | POP7 | 0.708086 | 13 | 0 | 12 |
| EPHB4 | SDC3 | 0.707411 | 4 | 0 | 4 |
For details and further investigation, click here