| Full name: inhibitor of DNA binding 1, HLH protein | Alias Symbol: dJ857M17.1.2|bHLHb24 | ||
| Type: protein-coding gene | Cytoband: 20q11.21 | ||
| Entrez ID: 3397 | HGNC ID: HGNC:5360 | Ensembl Gene: ENSG00000125968 | OMIM ID: 600349 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ID1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04015 | Rap1 signaling pathway | |
| hsa04390 | Hippo signaling pathway | |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells |
Expression of ID1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ID1 | 3397 | 208937_s_at | -0.6788 | 0.6428 | |
| GSE20347 | ID1 | 3397 | 208937_s_at | -0.4284 | 0.2495 | |
| GSE23400 | ID1 | 3397 | 208937_s_at | -0.1912 | 0.3428 | |
| GSE26886 | ID1 | 3397 | 208937_s_at | -0.9389 | 0.0204 | |
| GSE29001 | ID1 | 3397 | 208937_s_at | -0.7913 | 0.2232 | |
| GSE38129 | ID1 | 3397 | 208937_s_at | -0.2051 | 0.4413 | |
| GSE45670 | ID1 | 3397 | 208937_s_at | 0.4965 | 0.0568 | |
| GSE53622 | ID1 | 3397 | 86009 | -0.3565 | 0.0219 | |
| GSE53624 | ID1 | 3397 | 86009 | -0.1170 | 0.2662 | |
| GSE63941 | ID1 | 3397 | 208937_s_at | 1.0681 | 0.3094 | |
| GSE77861 | ID1 | 3397 | 208937_s_at | 0.8413 | 0.0154 | |
| GSE97050 | ID1 | 3397 | A_23_P252306 | 0.4820 | 0.5823 | |
| SRP007169 | ID1 | 3397 | RNAseq | -1.0936 | 0.0356 | |
| SRP008496 | ID1 | 3397 | RNAseq | -0.3956 | 0.4084 | |
| SRP064894 | ID1 | 3397 | RNAseq | -0.0843 | 0.7924 | |
| SRP133303 | ID1 | 3397 | RNAseq | 0.5554 | 0.1143 | |
| SRP159526 | ID1 | 3397 | RNAseq | -0.3375 | 0.4673 | |
| SRP193095 | ID1 | 3397 | RNAseq | -0.3035 | 0.1655 | |
| SRP219564 | ID1 | 3397 | RNAseq | -0.9556 | 0.0105 | |
| TCGA | ID1 | 3397 | RNAseq | 0.0218 | 0.8259 |
Upregulated datasets: 0; Downregulated datasets: 1.
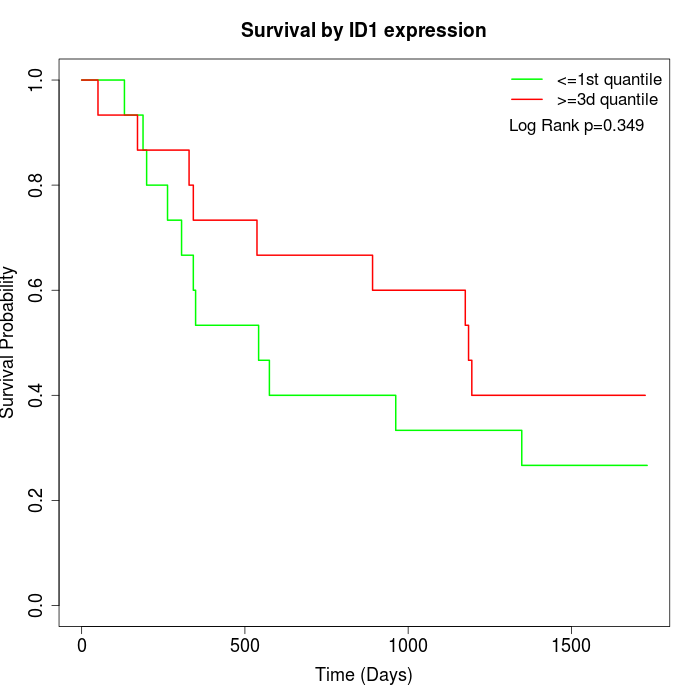
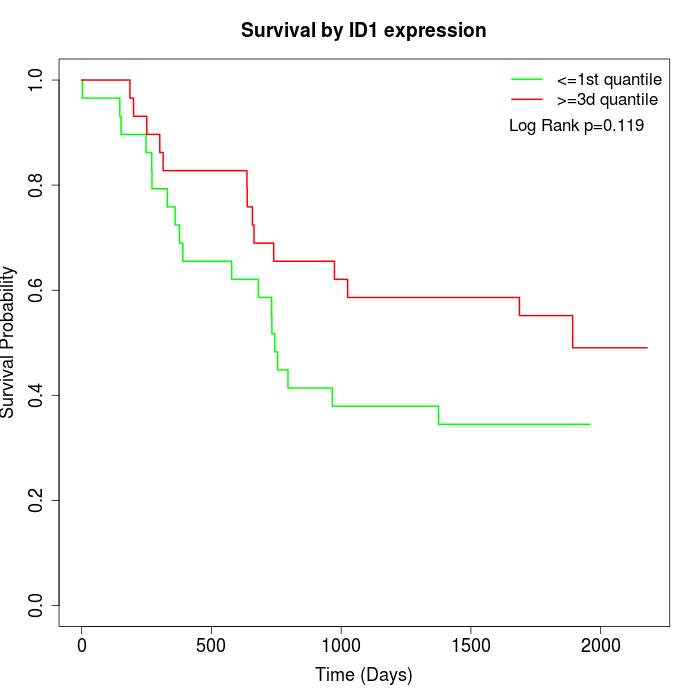
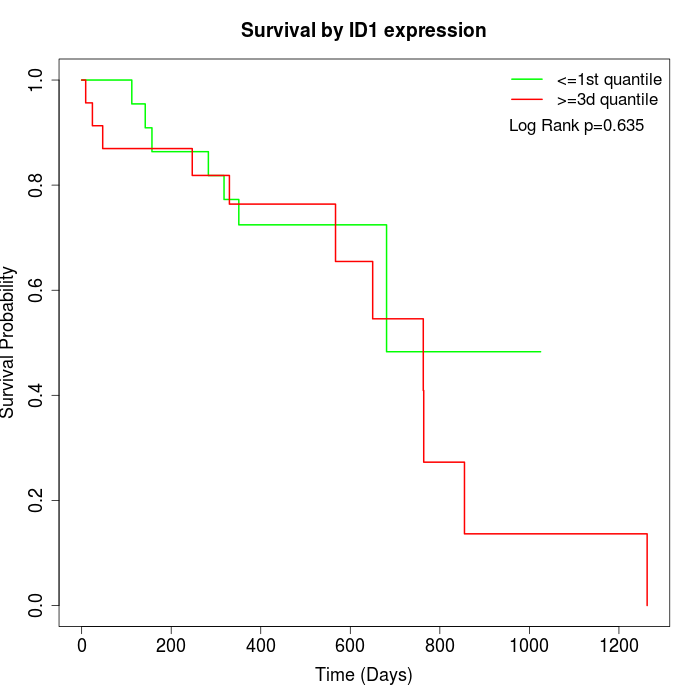
Survival by ID1 expression:
Note: Click image to view full size file.
Copy number change of ID1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ID1 | 3397 | 14 | 0 | 16 | |
| GSE20123 | ID1 | 3397 | 14 | 0 | 16 | |
| GSE43470 | ID1 | 3397 | 11 | 0 | 32 | |
| GSE46452 | ID1 | 3397 | 29 | 0 | 30 | |
| GSE47630 | ID1 | 3397 | 24 | 0 | 16 | |
| GSE54993 | ID1 | 3397 | 0 | 17 | 53 | |
| GSE54994 | ID1 | 3397 | 27 | 1 | 25 | |
| GSE60625 | ID1 | 3397 | 0 | 0 | 11 | |
| GSE74703 | ID1 | 3397 | 10 | 0 | 26 | |
| GSE74704 | ID1 | 3397 | 9 | 0 | 11 | |
| TCGA | ID1 | 3397 | 49 | 1 | 46 |
Total number of gains: 187; Total number of losses: 19; Total Number of normals: 282.
Somatic mutations of ID1:
Generating mutation plots.
Highly correlated genes for ID1:
Showing top 20/154 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ID1 | SNN | 0.769701 | 3 | 0 | 3 |
| ID1 | GOPC | 0.737393 | 3 | 0 | 3 |
| ID1 | FAM192A | 0.73442 | 3 | 0 | 3 |
| ID1 | TUFM | 0.716889 | 3 | 0 | 3 |
| ID1 | MBOAT2 | 0.711245 | 3 | 0 | 3 |
| ID1 | COASY | 0.7 | 3 | 0 | 3 |
| ID1 | SRXN1 | 0.699026 | 3 | 0 | 3 |
| ID1 | PLEKHG2 | 0.695545 | 3 | 0 | 3 |
| ID1 | DLX5 | 0.686859 | 3 | 0 | 3 |
| ID1 | SREBF2 | 0.682504 | 3 | 0 | 3 |
| ID1 | RAB8A | 0.679337 | 4 | 0 | 3 |
| ID1 | PSMD13 | 0.678926 | 3 | 0 | 3 |
| ID1 | NIT2 | 0.67547 | 3 | 0 | 3 |
| ID1 | EAF2 | 0.675016 | 3 | 0 | 3 |
| ID1 | EXOC6B | 0.670609 | 4 | 0 | 4 |
| ID1 | CDT1 | 0.669879 | 3 | 0 | 3 |
| ID1 | ARPC1A | 0.665727 | 4 | 0 | 3 |
| ID1 | PCK2 | 0.661326 | 3 | 0 | 3 |
| ID1 | CCNB2 | 0.660426 | 4 | 0 | 4 |
| ID1 | ID3 | 0.65723 | 4 | 0 | 4 |
For details and further investigation, click here