| Full name: cyclin B2 | Alias Symbol: HsT17299 | ||
| Type: protein-coding gene | Cytoband: 15q22.2 | ||
| Entrez ID: 9133 | HGNC ID: HGNC:1580 | Ensembl Gene: ENSG00000157456 | OMIM ID: 602755 |
| Drug and gene relationship at DGIdb | |||
CCNB2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04068 | FoxO signaling pathway | |
| hsa04110 | Cell cycle | |
| hsa04114 | Oocyte meiosis | |
| hsa04115 | p53 signaling pathway | |
| hsa05166 | HTLV-I infection |
Expression of CCNB2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCNB2 | 9133 | 202705_at | 0.1925 | 0.9283 | |
| GSE20347 | CCNB2 | 9133 | 202705_at | 1.5815 | 0.0000 | |
| GSE23400 | CCNB2 | 9133 | 202705_at | 0.9899 | 0.0000 | |
| GSE26886 | CCNB2 | 9133 | 202705_at | 1.1387 | 0.0025 | |
| GSE29001 | CCNB2 | 9133 | 202705_at | 1.3820 | 0.0019 | |
| GSE38129 | CCNB2 | 9133 | 202705_at | 1.9902 | 0.0000 | |
| GSE45670 | CCNB2 | 9133 | 202705_at | 1.3128 | 0.0000 | |
| GSE53622 | CCNB2 | 9133 | 81195 | 1.6437 | 0.0000 | |
| GSE53624 | CCNB2 | 9133 | 81195 | 1.9278 | 0.0000 | |
| GSE63941 | CCNB2 | 9133 | 202705_at | 3.3444 | 0.0001 | |
| GSE77861 | CCNB2 | 9133 | 202705_at | 0.8352 | 0.0038 | |
| GSE97050 | CCNB2 | 9133 | A_23_P65757 | 1.1114 | 0.0860 | |
| SRP007169 | CCNB2 | 9133 | RNAseq | 1.3978 | 0.0029 | |
| SRP008496 | CCNB2 | 9133 | RNAseq | 1.0654 | 0.0011 | |
| SRP064894 | CCNB2 | 9133 | RNAseq | 1.6250 | 0.0000 | |
| SRP133303 | CCNB2 | 9133 | RNAseq | 1.4213 | 0.0000 | |
| SRP159526 | CCNB2 | 9133 | RNAseq | 1.1840 | 0.0028 | |
| SRP193095 | CCNB2 | 9133 | RNAseq | 0.9024 | 0.0000 | |
| SRP219564 | CCNB2 | 9133 | RNAseq | 1.3100 | 0.0546 | |
| TCGA | CCNB2 | 9133 | RNAseq | 1.3448 | 0.0000 |
Upregulated datasets: 14; Downregulated datasets: 0.
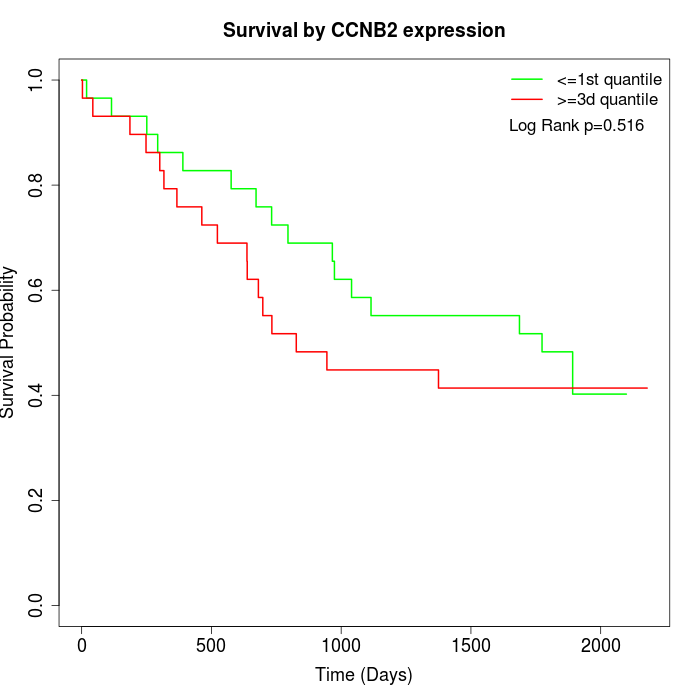
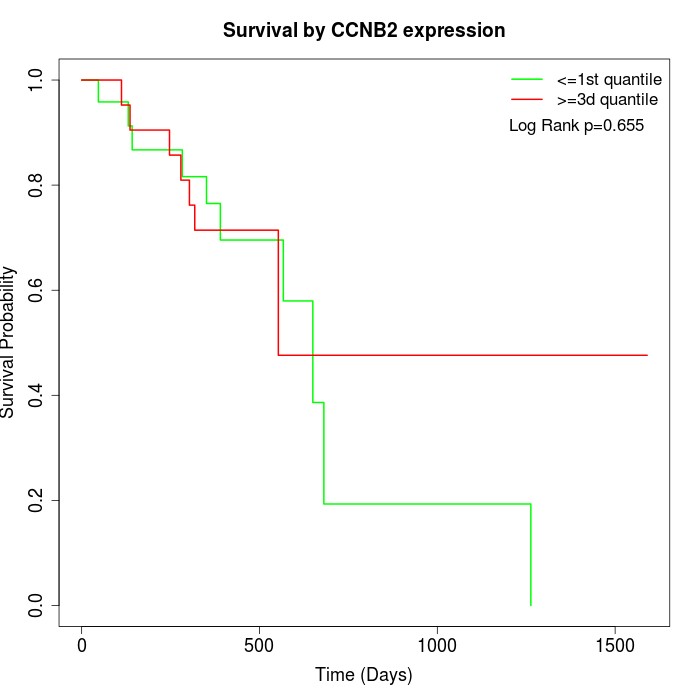
Survival by CCNB2 expression:
Note: Click image to view full size file.
Copy number change of CCNB2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCNB2 | 9133 | 6 | 4 | 20 | |
| GSE20123 | CCNB2 | 9133 | 6 | 3 | 21 | |
| GSE43470 | CCNB2 | 9133 | 4 | 4 | 35 | |
| GSE46452 | CCNB2 | 9133 | 3 | 7 | 49 | |
| GSE47630 | CCNB2 | 9133 | 8 | 10 | 22 | |
| GSE54993 | CCNB2 | 9133 | 5 | 6 | 59 | |
| GSE54994 | CCNB2 | 9133 | 6 | 8 | 39 | |
| GSE60625 | CCNB2 | 9133 | 4 | 0 | 7 | |
| GSE74703 | CCNB2 | 9133 | 4 | 3 | 29 | |
| GSE74704 | CCNB2 | 9133 | 2 | 3 | 15 | |
| TCGA | CCNB2 | 9133 | 10 | 16 | 70 |
Total number of gains: 58; Total number of losses: 64; Total Number of normals: 366.
Somatic mutations of CCNB2:
Generating mutation plots.
Highly correlated genes for CCNB2:
Showing top 20/1605 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCNB2 | UBE2C | 0.877561 | 12 | 0 | 12 |
| CCNB2 | NUSAP1 | 0.877246 | 12 | 0 | 12 |
| CCNB2 | BUB1B | 0.866922 | 9 | 0 | 8 |
| CCNB2 | KNSTRN | 0.861009 | 6 | 0 | 6 |
| CCNB2 | KIF23 | 0.859835 | 12 | 0 | 12 |
| CCNB2 | TOP2A | 0.856307 | 12 | 0 | 12 |
| CCNB2 | ASPM | 0.855476 | 12 | 0 | 12 |
| CCNB2 | TPX2 | 0.854543 | 12 | 0 | 12 |
| CCNB2 | RACGAP1 | 0.848766 | 10 | 0 | 10 |
| CCNB2 | SPAG5 | 0.847716 | 12 | 0 | 12 |
| CCNB2 | CDCA3 | 0.846251 | 12 | 0 | 12 |
| CCNB2 | GINS1 | 0.84606 | 11 | 0 | 11 |
| CCNB2 | CEP55 | 0.843913 | 12 | 0 | 12 |
| CCNB2 | TTK | 0.843576 | 12 | 0 | 11 |
| CCNB2 | DLGAP5 | 0.843536 | 11 | 0 | 11 |
| CCNB2 | NEK2 | 0.842586 | 12 | 0 | 12 |
| CCNB2 | PRC1 | 0.839109 | 13 | 0 | 12 |
| CCNB2 | CENPN | 0.836929 | 12 | 0 | 12 |
| CCNB2 | OIP5 | 0.835418 | 9 | 0 | 9 |
| CCNB2 | NUF2 | 0.835115 | 8 | 0 | 8 |
For details and further investigation, click here