| Full name: integrin subunit alpha D | Alias Symbol: CD11d|ADB2 | ||
| Type: protein-coding gene | Cytoband: 16p11.2 | ||
| Entrez ID: 3681 | HGNC ID: HGNC:6146 | Ensembl Gene: ENSG00000156886 | OMIM ID: 602453 |
| Drug and gene relationship at DGIdb | |||
ITGAD involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04810 | Regulation of actin cytoskeleton |
Expression of ITGAD:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ITGAD | 3681 | 1560686_at | -0.2227 | 0.4151 | |
| GSE26886 | ITGAD | 3681 | 1560686_at | -0.3946 | 0.0001 | |
| GSE45670 | ITGAD | 3681 | 1560686_at | -0.1681 | 0.1081 | |
| GSE53622 | ITGAD | 3681 | 115619 | 0.2241 | 0.1023 | |
| GSE53624 | ITGAD | 3681 | 115619 | -0.0163 | 0.9356 | |
| GSE63941 | ITGAD | 3681 | 1560686_at | 0.2372 | 0.0758 | |
| GSE77861 | ITGAD | 3681 | 1560686_at | -0.1771 | 0.0680 | |
| GSE97050 | ITGAD | 3681 | A_24_P366652 | 0.2566 | 0.3405 | |
| TCGA | ITGAD | 3681 | RNAseq | 0.7587 | 0.4970 |
Upregulated datasets: 0; Downregulated datasets: 0.
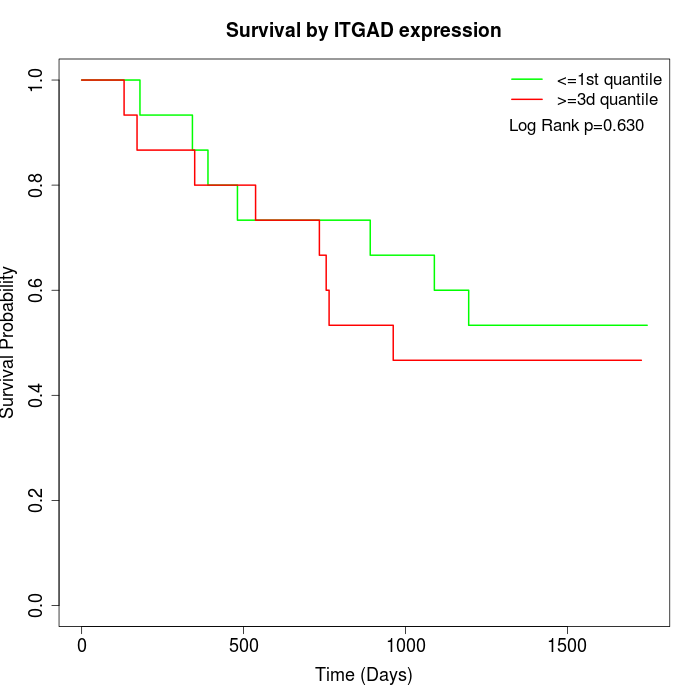
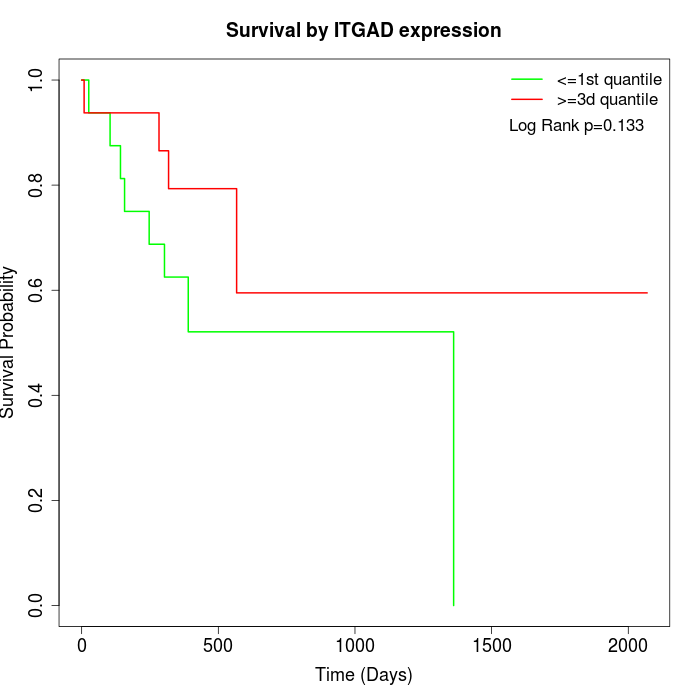
Survival by ITGAD expression:
Note: Click image to view full size file.
Copy number change of ITGAD:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ITGAD | 3681 | 4 | 5 | 21 | |
| GSE20123 | ITGAD | 3681 | 3 | 4 | 23 | |
| GSE43470 | ITGAD | 3681 | 3 | 3 | 37 | |
| GSE46452 | ITGAD | 3681 | 38 | 1 | 20 | |
| GSE47630 | ITGAD | 3681 | 11 | 7 | 22 | |
| GSE54993 | ITGAD | 3681 | 2 | 5 | 63 | |
| GSE54994 | ITGAD | 3681 | 4 | 10 | 39 | |
| GSE60625 | ITGAD | 3681 | 4 | 0 | 7 | |
| GSE74703 | ITGAD | 3681 | 3 | 2 | 31 | |
| GSE74704 | ITGAD | 3681 | 3 | 2 | 15 | |
| TCGA | ITGAD | 3681 | 20 | 10 | 66 |
Total number of gains: 95; Total number of losses: 49; Total Number of normals: 344.
Somatic mutations of ITGAD:
Generating mutation plots.
Highly correlated genes for ITGAD:
Showing top 20/528 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ITGAD | LCN2 | 0.761199 | 4 | 0 | 4 |
| ITGAD | RAB27A | 0.759753 | 3 | 0 | 3 |
| ITGAD | S100A12 | 0.748338 | 3 | 0 | 3 |
| ITGAD | PLEKHM1 | 0.729563 | 3 | 0 | 3 |
| ITGAD | TMPRSS11D | 0.724477 | 3 | 0 | 3 |
| ITGAD | RAB3D | 0.722862 | 3 | 0 | 3 |
| ITGAD | PI3 | 0.722674 | 3 | 0 | 3 |
| ITGAD | BBOX1 | 0.721021 | 4 | 0 | 4 |
| ITGAD | SERPINB4 | 0.716778 | 3 | 0 | 3 |
| ITGAD | SERPINB1 | 0.715464 | 4 | 0 | 4 |
| ITGAD | SLC46A2 | 0.71433 | 4 | 0 | 4 |
| ITGAD | IL22RA1 | 0.712605 | 3 | 0 | 3 |
| ITGAD | SPRR3 | 0.710113 | 3 | 0 | 3 |
| ITGAD | CXCR2 | 0.703654 | 4 | 0 | 4 |
| ITGAD | PDZK1IP1 | 0.702277 | 3 | 0 | 3 |
| ITGAD | TEX101 | 0.695885 | 4 | 0 | 3 |
| ITGAD | PALMD | 0.690273 | 4 | 0 | 4 |
| ITGAD | MAB21L3 | 0.687698 | 3 | 0 | 3 |
| ITGAD | SLC16A6 | 0.68731 | 4 | 0 | 4 |
| ITGAD | NLRX1 | 0.686424 | 4 | 0 | 4 |
For details and further investigation, click here