| Full name: NLR family pyrin domain containing 12 | Alias Symbol: RNO2|PYPAF7|Monarch1|PAN6|CLR19.3 | ||
| Type: protein-coding gene | Cytoband: 19q13.42 | ||
| Entrez ID: 91662 | HGNC ID: HGNC:22938 | Ensembl Gene: ENSG00000142405 | OMIM ID: 609648 |
| Drug and gene relationship at DGIdb | |||
Expression of NLRP12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NLRP12 | 91662 | 223944_at | -0.4179 | 0.0779 | |
| GSE26886 | NLRP12 | 91662 | 223944_at | -0.2591 | 0.0428 | |
| GSE45670 | NLRP12 | 91662 | 223944_at | -0.0501 | 0.6395 | |
| GSE53622 | NLRP12 | 91662 | 53019 | 0.2010 | 0.0502 | |
| GSE53624 | NLRP12 | 91662 | 53019 | 0.3589 | 0.0070 | |
| GSE63941 | NLRP12 | 91662 | 223944_at | 0.2691 | 0.0682 | |
| GSE77861 | NLRP12 | 91662 | 223944_at | -0.2585 | 0.0469 | |
| GSE97050 | NLRP12 | 91662 | A_23_P101434 | 0.0061 | 0.9837 | |
| SRP133303 | NLRP12 | 91662 | RNAseq | 0.2584 | 0.4528 | |
| SRP193095 | NLRP12 | 91662 | RNAseq | 0.5073 | 0.0368 | |
| TCGA | NLRP12 | 91662 | RNAseq | -0.3597 | 0.5503 |
Upregulated datasets: 0; Downregulated datasets: 0.
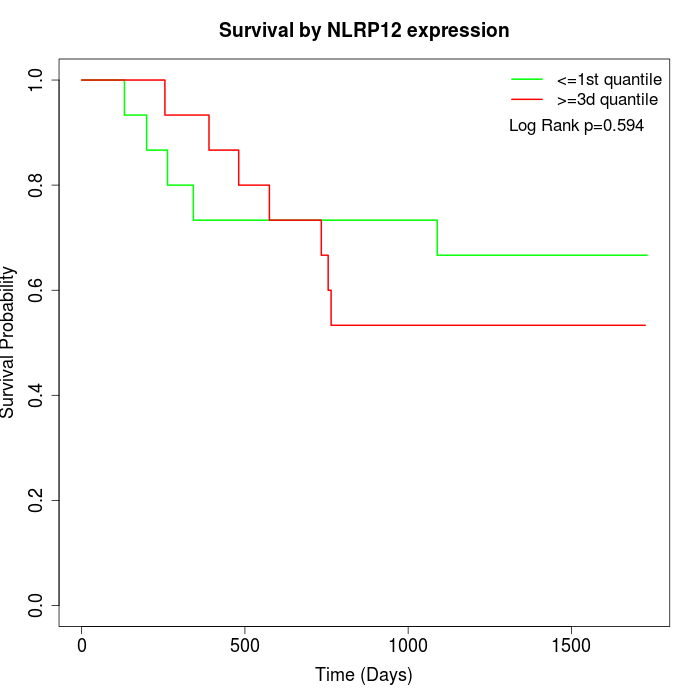
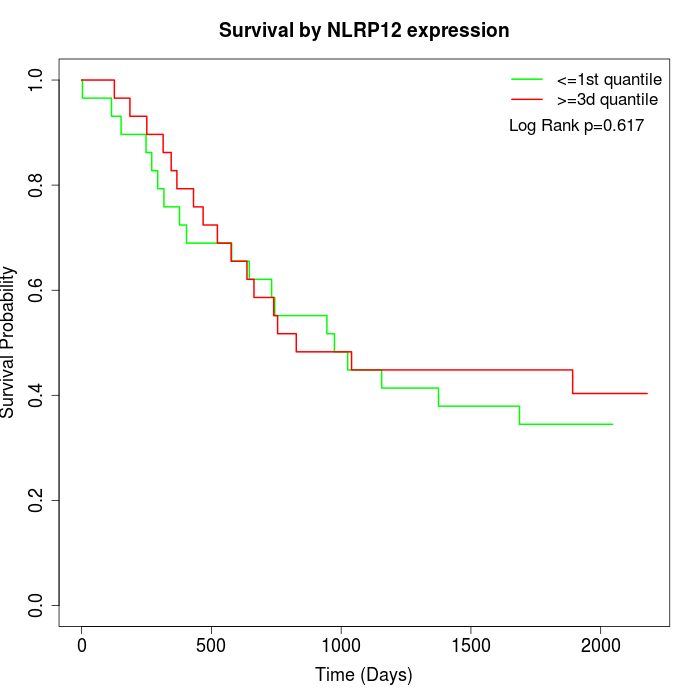
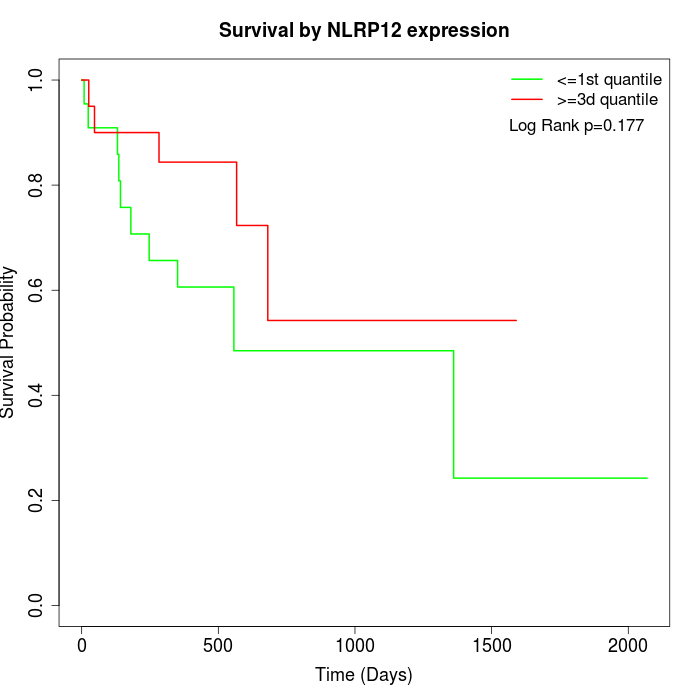
Survival by NLRP12 expression:
Note: Click image to view full size file.
Copy number change of NLRP12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NLRP12 | 91662 | 3 | 4 | 23 | |
| GSE20123 | NLRP12 | 91662 | 3 | 3 | 24 | |
| GSE43470 | NLRP12 | 91662 | 2 | 11 | 30 | |
| GSE46452 | NLRP12 | 91662 | 45 | 1 | 13 | |
| GSE47630 | NLRP12 | 91662 | 8 | 6 | 26 | |
| GSE54993 | NLRP12 | 91662 | 17 | 4 | 49 | |
| GSE54994 | NLRP12 | 91662 | 4 | 14 | 35 | |
| GSE60625 | NLRP12 | 91662 | 9 | 0 | 2 | |
| GSE74703 | NLRP12 | 91662 | 2 | 7 | 27 | |
| GSE74704 | NLRP12 | 91662 | 3 | 1 | 16 | |
| TCGA | NLRP12 | 91662 | 18 | 16 | 62 |
Total number of gains: 114; Total number of losses: 67; Total Number of normals: 307.
Somatic mutations of NLRP12:
Generating mutation plots.
Highly correlated genes for NLRP12:
Showing top 20/272 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NLRP12 | MYCT1 | 0.758742 | 3 | 0 | 3 |
| NLRP12 | CSDC2 | 0.7402 | 3 | 0 | 3 |
| NLRP12 | ADAMTS10 | 0.728278 | 3 | 0 | 3 |
| NLRP12 | TLE2 | 0.725832 | 3 | 0 | 3 |
| NLRP12 | SOX10 | 0.721744 | 5 | 0 | 5 |
| NLRP12 | TSSK2 | 0.718772 | 3 | 0 | 3 |
| NLRP12 | DEFB118 | 0.709134 | 4 | 0 | 4 |
| NLRP12 | CAST | 0.701009 | 3 | 0 | 3 |
| NLRP12 | BTD | 0.696852 | 3 | 0 | 3 |
| NLRP12 | CRYZL1 | 0.69528 | 3 | 0 | 3 |
| NLRP12 | KCTD6 | 0.68794 | 3 | 0 | 3 |
| NLRP12 | HDAC9 | 0.687287 | 3 | 0 | 3 |
| NLRP12 | RASSF3 | 0.684919 | 3 | 0 | 3 |
| NLRP12 | OXCT1 | 0.684453 | 3 | 0 | 3 |
| NLRP12 | ALG9 | 0.682005 | 3 | 0 | 3 |
| NLRP12 | SMARCA2 | 0.676452 | 3 | 0 | 3 |
| NLRP12 | GPS2 | 0.676108 | 3 | 0 | 3 |
| NLRP12 | SORBS2 | 0.675136 | 3 | 0 | 3 |
| NLRP12 | THSD4 | 0.674906 | 3 | 0 | 3 |
| NLRP12 | HTR3B | 0.673193 | 3 | 0 | 3 |
For details and further investigation, click here