| Full name: potassium two pore domain channel subfamily K member 13 | Alias Symbol: K2p13.1|THIK-1|THIK1 | ||
| Type: protein-coding gene | Cytoband: 14q32.11 | ||
| Entrez ID: 56659 | HGNC ID: HGNC:6275 | Ensembl Gene: ENSG00000152315 | OMIM ID: 607367 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNK13:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNK13 | 56659 | 221325_at | -0.0018 | 0.9966 | |
| GSE20347 | KCNK13 | 56659 | 221325_at | -0.1141 | 0.1260 | |
| GSE23400 | KCNK13 | 56659 | 221325_at | -0.0971 | 0.0054 | |
| GSE26886 | KCNK13 | 56659 | 221325_at | 0.0673 | 0.6249 | |
| GSE29001 | KCNK13 | 56659 | 221325_at | -0.1543 | 0.1891 | |
| GSE38129 | KCNK13 | 56659 | 221325_at | -0.1255 | 0.0306 | |
| GSE45670 | KCNK13 | 56659 | 221325_at | 0.0675 | 0.3850 | |
| GSE53622 | KCNK13 | 56659 | 24700 | 0.5814 | 0.0004 | |
| GSE53624 | KCNK13 | 56659 | 24700 | 0.6260 | 0.0007 | |
| GSE63941 | KCNK13 | 56659 | 221325_at | -0.1213 | 0.4151 | |
| GSE77861 | KCNK13 | 56659 | 221325_at | -0.2114 | 0.0264 | |
| GSE97050 | KCNK13 | 56659 | A_23_P3177 | 0.0555 | 0.8076 | |
| SRP133303 | KCNK13 | 56659 | RNAseq | 1.5315 | 0.0002 | |
| TCGA | KCNK13 | 56659 | RNAseq | 1.6536 | 0.0000 |
Upregulated datasets: 2; Downregulated datasets: 0.
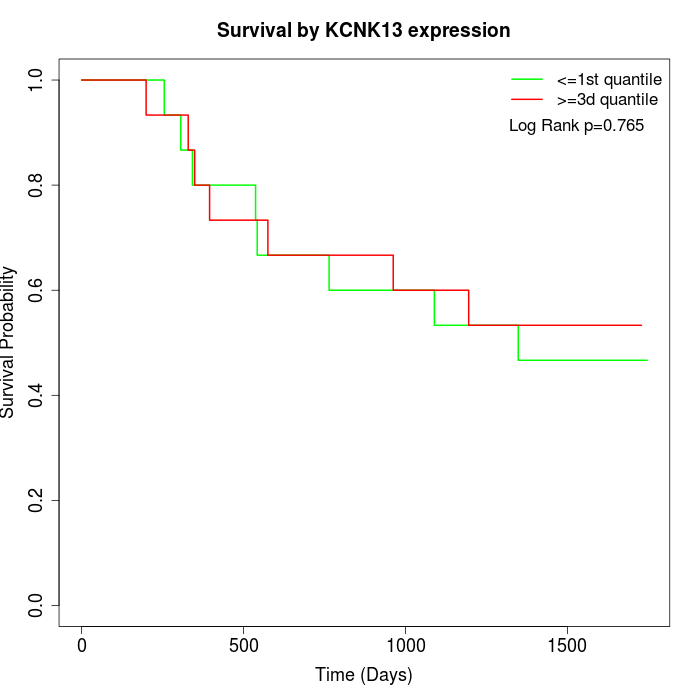
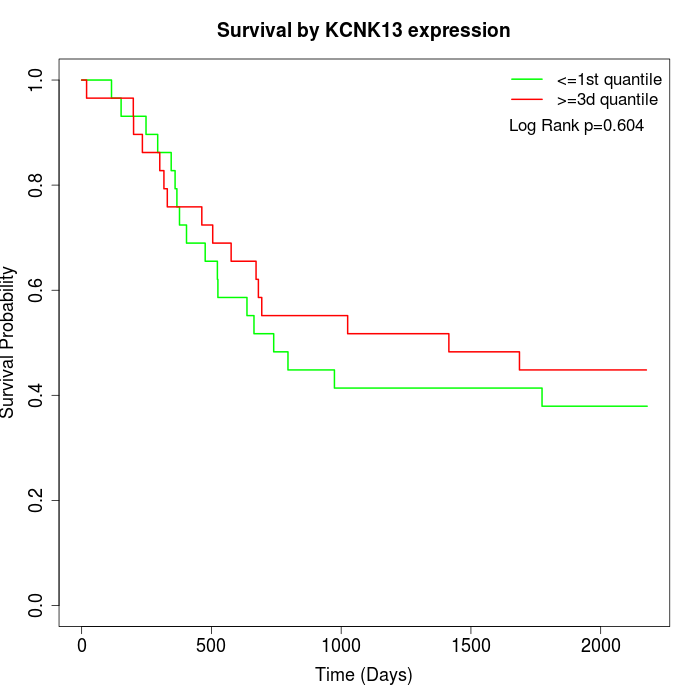
Survival by KCNK13 expression:
Note: Click image to view full size file.
Copy number change of KCNK13:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNK13 | 56659 | 9 | 5 | 16 | |
| GSE20123 | KCNK13 | 56659 | 9 | 5 | 16 | |
| GSE43470 | KCNK13 | 56659 | 7 | 3 | 33 | |
| GSE46452 | KCNK13 | 56659 | 16 | 3 | 40 | |
| GSE47630 | KCNK13 | 56659 | 11 | 8 | 21 | |
| GSE54993 | KCNK13 | 56659 | 3 | 8 | 59 | |
| GSE54994 | KCNK13 | 56659 | 19 | 4 | 30 | |
| GSE60625 | KCNK13 | 56659 | 0 | 2 | 9 | |
| GSE74703 | KCNK13 | 56659 | 6 | 3 | 27 | |
| GSE74704 | KCNK13 | 56659 | 4 | 4 | 12 | |
| TCGA | KCNK13 | 56659 | 32 | 20 | 44 |
Total number of gains: 116; Total number of losses: 65; Total Number of normals: 307.
Somatic mutations of KCNK13:
Generating mutation plots.
Highly correlated genes for KCNK13:
Showing top 20/408 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNK13 | VPS52 | 0.743518 | 3 | 0 | 3 |
| KCNK13 | EPN1 | 0.721536 | 3 | 0 | 3 |
| KCNK13 | PPIC | 0.717814 | 3 | 0 | 3 |
| KCNK13 | CHRNB1 | 0.708878 | 3 | 0 | 3 |
| KCNK13 | ALG14 | 0.704486 | 3 | 0 | 3 |
| KCNK13 | NDUFC1 | 0.699916 | 3 | 0 | 3 |
| KCNK13 | NR5A2 | 0.685714 | 3 | 0 | 3 |
| KCNK13 | ARHGEF6 | 0.681565 | 3 | 0 | 3 |
| KCNK13 | YOD1 | 0.669089 | 3 | 0 | 3 |
| KCNK13 | CNR2 | 0.667086 | 6 | 0 | 6 |
| KCNK13 | RABEP2 | 0.665492 | 4 | 0 | 3 |
| KCNK13 | EPN3 | 0.657998 | 3 | 0 | 3 |
| KCNK13 | OSBPL10 | 0.657564 | 4 | 0 | 3 |
| KCNK13 | ARL6IP5 | 0.657264 | 3 | 0 | 3 |
| KCNK13 | ITPRIP | 0.655442 | 3 | 0 | 3 |
| KCNK13 | FUT9 | 0.655332 | 4 | 0 | 4 |
| KCNK13 | GYG2 | 0.654902 | 6 | 0 | 6 |
| KCNK13 | RPLP2 | 0.653742 | 3 | 0 | 3 |
| KCNK13 | PF4 | 0.653392 | 3 | 0 | 3 |
| KCNK13 | IFNA16 | 0.65217 | 3 | 0 | 3 |
For details and further investigation, click here