| Full name: potassium calcium-activated channel subfamily N member 2 | Alias Symbol: KCa2.2|hSK2 | ||
| Type: protein-coding gene | Cytoband: 5q22.3 | ||
| Entrez ID: 3781 | HGNC ID: HGNC:6291 | Ensembl Gene: ENSG00000080709 | OMIM ID: 605879 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of KCNN2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNN2 | 3781 | 220116_at | -0.1440 | 0.9204 | |
| GSE20347 | KCNN2 | 3781 | 220116_at | 0.0428 | 0.5484 | |
| GSE23400 | KCNN2 | 3781 | 220116_at | -0.0496 | 0.0735 | |
| GSE26886 | KCNN2 | 3781 | 220116_at | 0.0945 | 0.3518 | |
| GSE29001 | KCNN2 | 3781 | 220116_at | -0.0310 | 0.8800 | |
| GSE38129 | KCNN2 | 3781 | 220116_at | -0.1672 | 0.4322 | |
| GSE45670 | KCNN2 | 3781 | 220116_at | -0.0238 | 0.9081 | |
| GSE53622 | KCNN2 | 3781 | 36629 | -0.2235 | 0.0213 | |
| GSE53624 | KCNN2 | 3781 | 36629 | -0.3024 | 0.0044 | |
| GSE63941 | KCNN2 | 3781 | 220116_at | -0.0713 | 0.8889 | |
| GSE77861 | KCNN2 | 3781 | 220116_at | -0.0221 | 0.7672 | |
| GSE97050 | KCNN2 | 3781 | A_23_P500353 | -0.2910 | 0.1848 | |
| SRP133303 | KCNN2 | 3781 | RNAseq | 0.5115 | 0.3130 | |
| SRP159526 | KCNN2 | 3781 | RNAseq | 1.1946 | 0.0381 | |
| TCGA | KCNN2 | 3781 | RNAseq | -0.3045 | 0.3955 |
Upregulated datasets: 1; Downregulated datasets: 0.
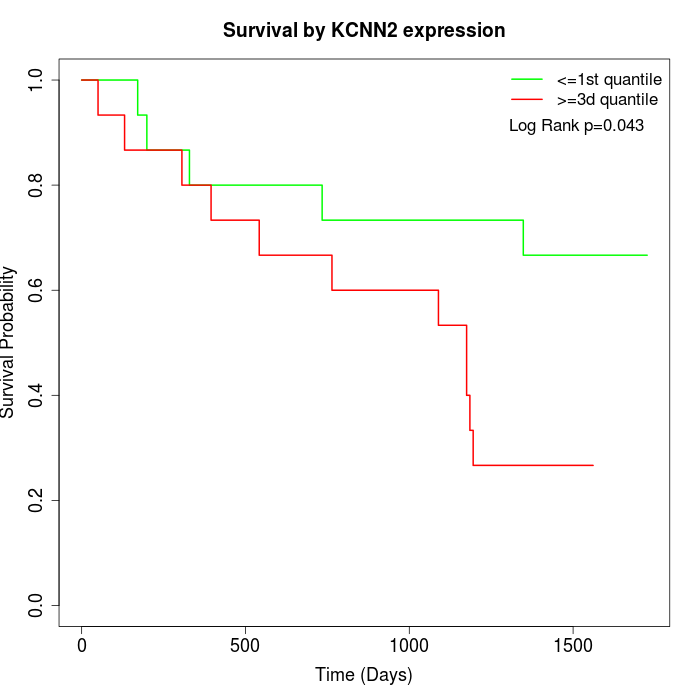
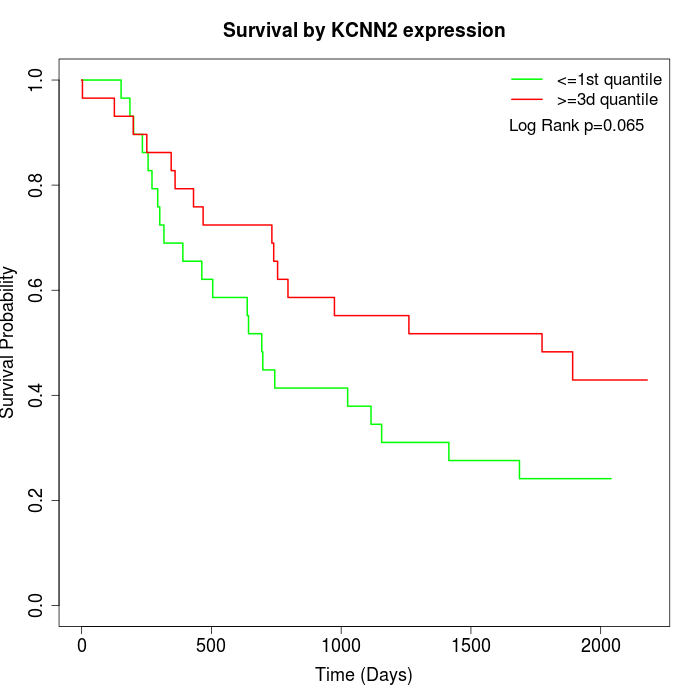
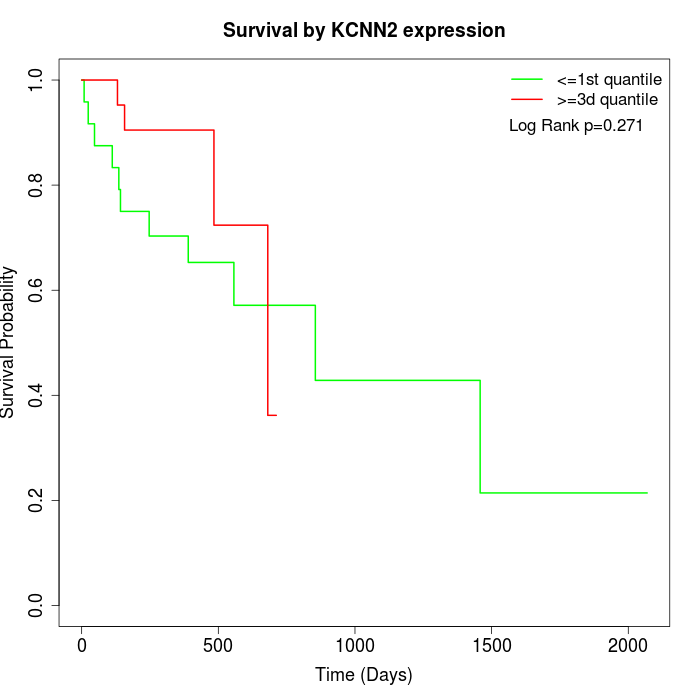
Survival by KCNN2 expression:
Note: Click image to view full size file.
Copy number change of KCNN2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNN2 | 3781 | 1 | 13 | 16 | |
| GSE20123 | KCNN2 | 3781 | 1 | 13 | 16 | |
| GSE43470 | KCNN2 | 3781 | 3 | 8 | 32 | |
| GSE46452 | KCNN2 | 3781 | 0 | 28 | 31 | |
| GSE47630 | KCNN2 | 3781 | 0 | 21 | 19 | |
| GSE54993 | KCNN2 | 3781 | 9 | 1 | 60 | |
| GSE54994 | KCNN2 | 3781 | 1 | 15 | 37 | |
| GSE60625 | KCNN2 | 3781 | 0 | 0 | 11 | |
| GSE74703 | KCNN2 | 3781 | 2 | 6 | 28 | |
| GSE74704 | KCNN2 | 3781 | 1 | 7 | 12 | |
| TCGA | KCNN2 | 3781 | 2 | 41 | 53 |
Total number of gains: 20; Total number of losses: 153; Total Number of normals: 315.
Somatic mutations of KCNN2:
Generating mutation plots.
Highly correlated genes for KCNN2:
Showing top 20/175 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNN2 | TSPAN2 | 0.693097 | 3 | 0 | 3 |
| KCNN2 | NPY | 0.687123 | 5 | 0 | 5 |
| KCNN2 | CCDC8 | 0.673176 | 3 | 0 | 3 |
| KCNN2 | PLCL1 | 0.670837 | 5 | 0 | 5 |
| KCNN2 | SUPT6H | 0.666625 | 3 | 0 | 3 |
| KCNN2 | IQSEC1 | 0.65876 | 3 | 0 | 3 |
| KCNN2 | CLCNKA | 0.656629 | 3 | 0 | 3 |
| KCNN2 | SYPL2 | 0.656434 | 3 | 0 | 3 |
| KCNN2 | CARTPT | 0.650598 | 5 | 0 | 4 |
| KCNN2 | SHISA8 | 0.646735 | 4 | 0 | 3 |
| KCNN2 | MYO16 | 0.646137 | 3 | 0 | 3 |
| KCNN2 | PRR18 | 0.637279 | 3 | 0 | 3 |
| KCNN2 | BEST4 | 0.633586 | 3 | 0 | 3 |
| KCNN2 | PEBP4 | 0.632503 | 3 | 0 | 3 |
| KCNN2 | PLA2G12B | 0.632203 | 4 | 0 | 3 |
| KCNN2 | BEX1 | 0.63114 | 3 | 0 | 3 |
| KCNN2 | KLHL26 | 0.631059 | 4 | 0 | 3 |
| KCNN2 | FOXA3 | 0.622097 | 5 | 0 | 4 |
| KCNN2 | C2CD4C | 0.621497 | 3 | 0 | 3 |
| KCNN2 | KATNAL2 | 0.618811 | 3 | 0 | 3 |
For details and further investigation, click here