| Full name: potassium voltage-gated channel subfamily Q member 5 | Alias Symbol: Kv7.5 | ||
| Type: protein-coding gene | Cytoband: 6q13 | ||
| Entrez ID: 56479 | HGNC ID: HGNC:6299 | Ensembl Gene: ENSG00000185760 | OMIM ID: 607357 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of KCNQ5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNQ5 | 56479 | 244623_at | 0.3724 | 0.5465 | |
| GSE26886 | KCNQ5 | 56479 | 244623_at | 1.1156 | 0.0017 | |
| GSE45670 | KCNQ5 | 56479 | 244623_at | 0.6195 | 0.1143 | |
| GSE53622 | KCNQ5 | 56479 | 133226 | 1.0039 | 0.0000 | |
| GSE53624 | KCNQ5 | 56479 | 133226 | 0.7604 | 0.0000 | |
| GSE63941 | KCNQ5 | 56479 | 244623_at | -0.0303 | 0.9790 | |
| GSE77861 | KCNQ5 | 56479 | 244623_at | 0.3612 | 0.0953 | |
| GSE97050 | KCNQ5 | 56479 | A_23_P167841 | 0.5462 | 0.1785 | |
| SRP007169 | KCNQ5 | 56479 | RNAseq | 3.1410 | 0.0009 | |
| SRP133303 | KCNQ5 | 56479 | RNAseq | 1.0504 | 0.0070 | |
| SRP159526 | KCNQ5 | 56479 | RNAseq | 1.1537 | 0.0287 | |
| SRP193095 | KCNQ5 | 56479 | RNAseq | 1.2147 | 0.0000 | |
| SRP219564 | KCNQ5 | 56479 | RNAseq | 1.3192 | 0.1209 | |
| TCGA | KCNQ5 | 56479 | RNAseq | 0.3001 | 0.2257 |
Upregulated datasets: 6; Downregulated datasets: 0.
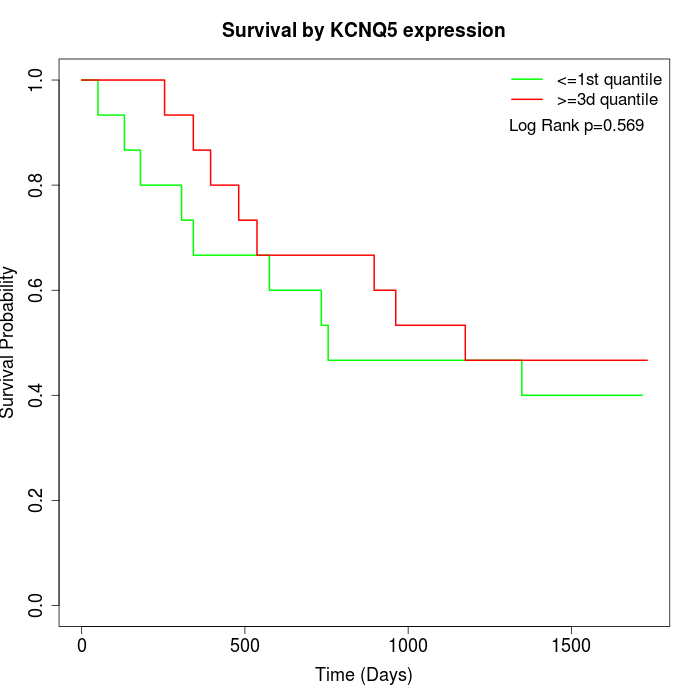
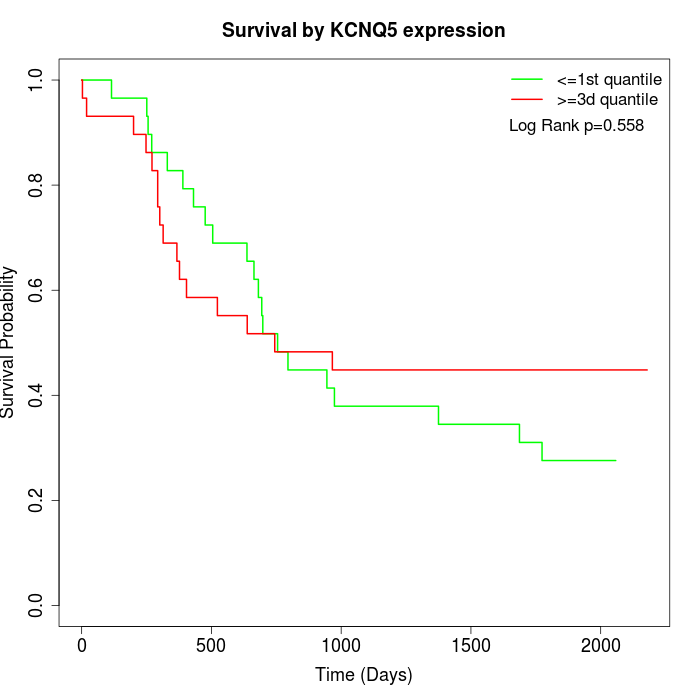
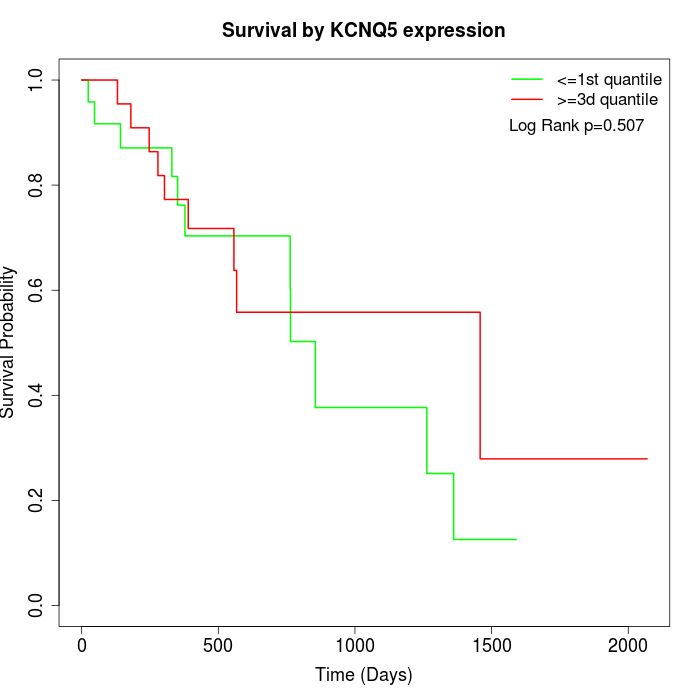
Survival by KCNQ5 expression:
Note: Click image to view full size file.
Copy number change of KCNQ5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNQ5 | 56479 | 1 | 3 | 26 | |
| GSE20123 | KCNQ5 | 56479 | 1 | 2 | 27 | |
| GSE43470 | KCNQ5 | 56479 | 3 | 0 | 40 | |
| GSE46452 | KCNQ5 | 56479 | 2 | 11 | 46 | |
| GSE47630 | KCNQ5 | 56479 | 8 | 4 | 28 | |
| GSE54993 | KCNQ5 | 56479 | 3 | 2 | 65 | |
| GSE54994 | KCNQ5 | 56479 | 9 | 5 | 39 | |
| GSE60625 | KCNQ5 | 56479 | 0 | 1 | 10 | |
| GSE74703 | KCNQ5 | 56479 | 3 | 0 | 33 | |
| GSE74704 | KCNQ5 | 56479 | 0 | 1 | 19 | |
| TCGA | KCNQ5 | 56479 | 9 | 21 | 66 |
Total number of gains: 39; Total number of losses: 50; Total Number of normals: 399.
Somatic mutations of KCNQ5:
Generating mutation plots.
Highly correlated genes for KCNQ5:
Showing top 20/242 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNQ5 | CYP2S1 | 0.732297 | 3 | 0 | 3 |
| KCNQ5 | FGF19 | 0.708301 | 3 | 0 | 3 |
| KCNQ5 | SLC12A7 | 0.705569 | 3 | 0 | 3 |
| KCNQ5 | RHPN2 | 0.671884 | 4 | 0 | 4 |
| KCNQ5 | ARSJ | 0.66999 | 3 | 0 | 3 |
| KCNQ5 | PLEKHG4B | 0.666065 | 3 | 0 | 3 |
| KCNQ5 | MUTYH | 0.663456 | 4 | 0 | 3 |
| KCNQ5 | SLCO1B3 | 0.658223 | 4 | 0 | 4 |
| KCNQ5 | SH3BP4 | 0.647619 | 5 | 0 | 4 |
| KCNQ5 | GDF15 | 0.643319 | 3 | 0 | 3 |
| KCNQ5 | MYO10 | 0.631857 | 3 | 0 | 3 |
| KCNQ5 | PRSS21 | 0.626678 | 5 | 0 | 3 |
| KCNQ5 | SLC38A10 | 0.624168 | 4 | 0 | 4 |
| KCNQ5 | POLE | 0.624116 | 5 | 0 | 4 |
| KCNQ5 | PPP1R35 | 0.621245 | 4 | 0 | 3 |
| KCNQ5 | FAM86C1 | 0.620322 | 3 | 0 | 3 |
| KCNQ5 | NIPA2 | 0.6124 | 4 | 0 | 3 |
| KCNQ5 | CDC6 | 0.611376 | 4 | 0 | 3 |
| KCNQ5 | CTDSPL2 | 0.60657 | 4 | 0 | 4 |
| KCNQ5 | VPS37D | 0.605936 | 3 | 0 | 3 |
For details and further investigation, click here