| Full name: liver enriched antimicrobial peptide 2 | Alias Symbol: LEAP-2 | ||
| Type: protein-coding gene | Cytoband: 5q31.1 | ||
| Entrez ID: 116842 | HGNC ID: HGNC:29571 | Ensembl Gene: ENSG00000164406 | OMIM ID: 611373 |
| Drug and gene relationship at DGIdb | |||
Expression of LEAP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LEAP2 | 116842 | 1552362_a_at | -0.2432 | 0.5263 | |
| GSE26886 | LEAP2 | 116842 | 1552362_a_at | -0.7265 | 0.0079 | |
| GSE45670 | LEAP2 | 116842 | 1552362_a_at | -0.1007 | 0.7428 | |
| GSE53622 | LEAP2 | 116842 | 64000 | -0.5890 | 0.0000 | |
| GSE53624 | LEAP2 | 116842 | 9111 | -0.2493 | 0.0053 | |
| GSE63941 | LEAP2 | 116842 | 1552362_a_at | 0.9164 | 0.0457 | |
| GSE77861 | LEAP2 | 116842 | 1552362_a_at | -0.0602 | 0.4943 | |
| GSE97050 | LEAP2 | 116842 | A_33_P3341259 | -0.6996 | 0.1236 | |
| SRP133303 | LEAP2 | 116842 | RNAseq | -0.9681 | 0.0000 | |
| SRP159526 | LEAP2 | 116842 | RNAseq | -0.0343 | 0.9539 | |
| SRP193095 | LEAP2 | 116842 | RNAseq | -0.8848 | 0.0000 | |
| SRP219564 | LEAP2 | 116842 | RNAseq | -0.3618 | 0.4044 | |
| TCGA | LEAP2 | 116842 | RNAseq | -1.2000 | 0.0005 |
Upregulated datasets: 0; Downregulated datasets: 1.
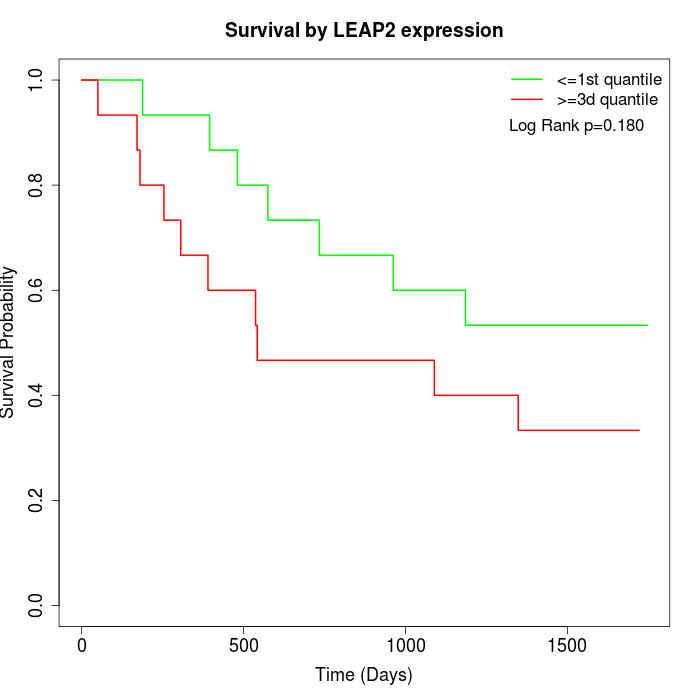
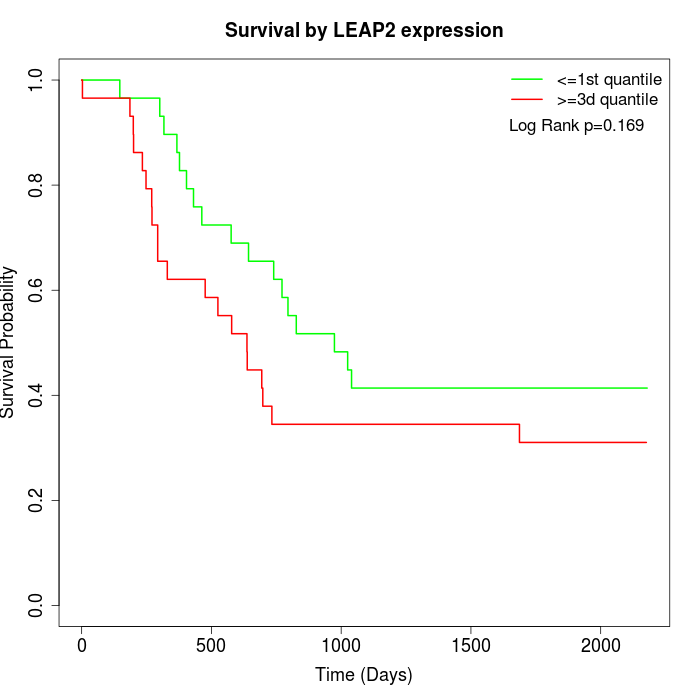
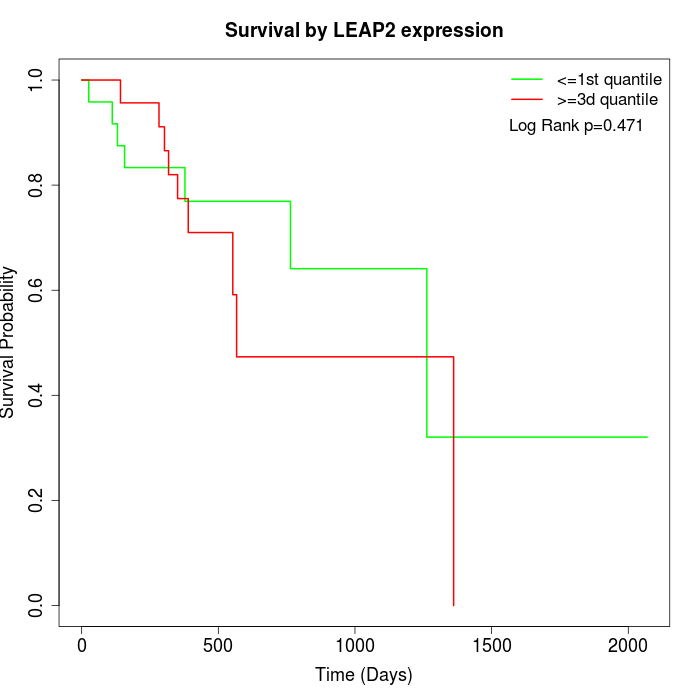
Survival by LEAP2 expression:
Note: Click image to view full size file.
Copy number change of LEAP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LEAP2 | 116842 | 1 | 11 | 18 | |
| GSE20123 | LEAP2 | 116842 | 2 | 11 | 17 | |
| GSE43470 | LEAP2 | 116842 | 2 | 9 | 32 | |
| GSE46452 | LEAP2 | 116842 | 0 | 27 | 32 | |
| GSE47630 | LEAP2 | 116842 | 0 | 21 | 19 | |
| GSE54993 | LEAP2 | 116842 | 9 | 1 | 60 | |
| GSE54994 | LEAP2 | 116842 | 2 | 14 | 37 | |
| GSE60625 | LEAP2 | 116842 | 0 | 0 | 11 | |
| GSE74703 | LEAP2 | 116842 | 2 | 6 | 28 | |
| GSE74704 | LEAP2 | 116842 | 1 | 5 | 14 | |
| TCGA | LEAP2 | 116842 | 3 | 39 | 54 |
Total number of gains: 22; Total number of losses: 144; Total Number of normals: 322.
Somatic mutations of LEAP2:
Generating mutation plots.
Highly correlated genes for LEAP2:
Showing top 20/39 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LEAP2 | ACACB | 0.672005 | 3 | 0 | 3 |
| LEAP2 | PPP1CB | 0.666254 | 3 | 0 | 3 |
| LEAP2 | BTC | 0.634873 | 3 | 0 | 3 |
| LEAP2 | RAD23B | 0.634248 | 3 | 0 | 3 |
| LEAP2 | STX19 | 0.630175 | 3 | 0 | 3 |
| LEAP2 | ANK3 | 0.629132 | 4 | 0 | 4 |
| LEAP2 | AIF1L | 0.623167 | 3 | 0 | 3 |
| LEAP2 | MYL5 | 0.617681 | 4 | 0 | 3 |
| LEAP2 | PYGM | 0.615384 | 3 | 0 | 3 |
| LEAP2 | RMND5A | 0.614285 | 4 | 0 | 3 |
| LEAP2 | MPZL3 | 0.610313 | 3 | 0 | 3 |
| LEAP2 | TMEM125 | 0.610112 | 3 | 0 | 3 |
| LEAP2 | DCAF8 | 0.609193 | 4 | 0 | 3 |
| LEAP2 | NDUFV3 | 0.601903 | 3 | 0 | 3 |
| LEAP2 | ADCYAP1R1 | 0.595404 | 3 | 0 | 3 |
| LEAP2 | C1orf56 | 0.593831 | 3 | 0 | 3 |
| LEAP2 | MYOT | 0.593193 | 3 | 0 | 3 |
| LEAP2 | NEBL | 0.590653 | 3 | 0 | 3 |
| LEAP2 | PPARGC1A | 0.589236 | 3 | 0 | 3 |
| LEAP2 | GABPA | 0.588782 | 3 | 0 | 3 |
For details and further investigation, click here