| Full name: legumain | Alias Symbol: LGMN1 | ||
| Type: protein-coding gene | Cytoband: 14q32.12 | ||
| Entrez ID: 5641 | HGNC ID: HGNC:9472 | Ensembl Gene: ENSG00000100600 | OMIM ID: 602620 |
| Drug and gene relationship at DGIdb | |||
LGMN involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04612 | Antigen processing and presentation |
Expression of LGMN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LGMN | 5641 | 201212_at | -0.1728 | 0.7734 | |
| GSE20347 | LGMN | 5641 | 201212_at | -0.0229 | 0.9230 | |
| GSE23400 | LGMN | 5641 | 201212_at | 0.0846 | 0.3753 | |
| GSE26886 | LGMN | 5641 | 201212_at | -0.4655 | 0.0026 | |
| GSE29001 | LGMN | 5641 | 201212_at | -0.1678 | 0.6571 | |
| GSE38129 | LGMN | 5641 | 201212_at | 0.0165 | 0.9365 | |
| GSE45670 | LGMN | 5641 | 201212_at | -0.2248 | 0.2535 | |
| GSE53622 | LGMN | 5641 | 86778 | 0.0338 | 0.6916 | |
| GSE53624 | LGMN | 5641 | 86778 | 0.0648 | 0.3015 | |
| GSE63941 | LGMN | 5641 | 201212_at | -0.0455 | 0.9344 | |
| GSE77861 | LGMN | 5641 | 201212_at | -0.1727 | 0.4742 | |
| GSE97050 | LGMN | 5641 | A_23_P25994 | 0.6474 | 0.1702 | |
| SRP007169 | LGMN | 5641 | RNAseq | 0.5664 | 0.3194 | |
| SRP008496 | LGMN | 5641 | RNAseq | 0.7995 | 0.0387 | |
| SRP064894 | LGMN | 5641 | RNAseq | 0.3737 | 0.0959 | |
| SRP133303 | LGMN | 5641 | RNAseq | 0.3608 | 0.1245 | |
| SRP159526 | LGMN | 5641 | RNAseq | 0.3213 | 0.3231 | |
| SRP193095 | LGMN | 5641 | RNAseq | -0.1013 | 0.5748 | |
| SRP219564 | LGMN | 5641 | RNAseq | 0.8347 | 0.1439 | |
| TCGA | LGMN | 5641 | RNAseq | -0.1162 | 0.0414 |
Upregulated datasets: 0; Downregulated datasets: 0.
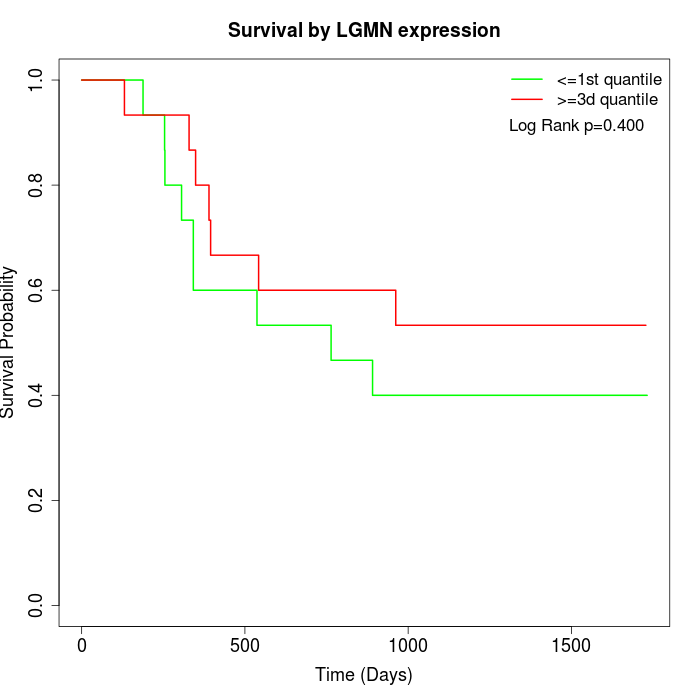
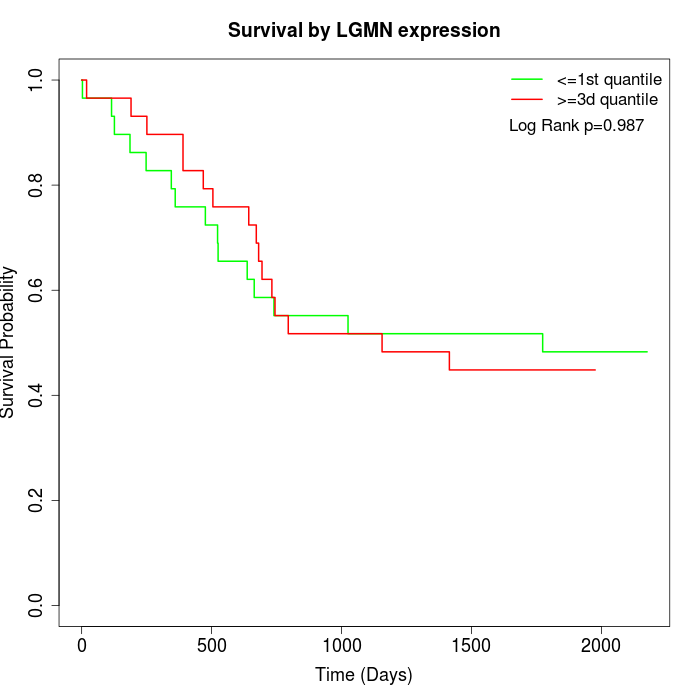
Survival by LGMN expression:
Note: Click image to view full size file.
Copy number change of LGMN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LGMN | 5641 | 8 | 5 | 17 | |
| GSE20123 | LGMN | 5641 | 8 | 4 | 18 | |
| GSE43470 | LGMN | 5641 | 7 | 3 | 33 | |
| GSE46452 | LGMN | 5641 | 16 | 3 | 40 | |
| GSE47630 | LGMN | 5641 | 11 | 8 | 21 | |
| GSE54993 | LGMN | 5641 | 3 | 8 | 59 | |
| GSE54994 | LGMN | 5641 | 20 | 4 | 29 | |
| GSE60625 | LGMN | 5641 | 0 | 2 | 9 | |
| GSE74703 | LGMN | 5641 | 6 | 3 | 27 | |
| GSE74704 | LGMN | 5641 | 4 | 3 | 13 | |
| TCGA | LGMN | 5641 | 29 | 21 | 46 |
Total number of gains: 112; Total number of losses: 64; Total Number of normals: 312.
Somatic mutations of LGMN:
Generating mutation plots.
Highly correlated genes for LGMN:
Showing top 20/70 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LGMN | ANKRD37 | 0.702572 | 3 | 0 | 3 |
| LGMN | HILPDA | 0.671522 | 3 | 0 | 3 |
| LGMN | ASAH1 | 0.670784 | 3 | 0 | 3 |
| LGMN | METTL9 | 0.668996 | 3 | 0 | 3 |
| LGMN | CYP4F22 | 0.634603 | 3 | 0 | 3 |
| LGMN | NABP1 | 0.631641 | 3 | 0 | 3 |
| LGMN | MBD2 | 0.626801 | 3 | 0 | 3 |
| LGMN | TM9SF2 | 0.622893 | 4 | 0 | 3 |
| LGMN | CITED2 | 0.61511 | 3 | 0 | 3 |
| LGMN | RCN3 | 0.611129 | 4 | 0 | 4 |
| LGMN | DUSP11 | 0.604926 | 4 | 0 | 4 |
| LGMN | LPIN2 | 0.594451 | 4 | 0 | 3 |
| LGMN | TMEM59 | 0.593389 | 4 | 0 | 4 |
| LGMN | GDI2 | 0.59271 | 4 | 0 | 3 |
| LGMN | PEX3 | 0.588189 | 4 | 0 | 3 |
| LGMN | ATP6V1C2 | 0.58377 | 4 | 0 | 3 |
| LGMN | C1GALT1 | 0.582446 | 3 | 0 | 3 |
| LGMN | CAB39 | 0.580977 | 4 | 0 | 3 |
| LGMN | ASL | 0.577487 | 3 | 0 | 3 |
| LGMN | SCYL2 | 0.577328 | 4 | 0 | 3 |
For details and further investigation, click here