| Full name: LLGL scribble cell polarity complex component 2 | Alias Symbol: HGL|Hugl-2 | ||
| Type: protein-coding gene | Cytoband: 17q25.1 | ||
| Entrez ID: 3993 | HGNC ID: HGNC:6629 | Ensembl Gene: ENSG00000073350 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
LLGL2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04390 | Hippo signaling pathway | |
| hsa04530 | Tight junction |
Expression of LLGL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LLGL2 | 3993 | 1554006_a_at | -0.4595 | 0.4847 | |
| GSE20347 | LLGL2 | 3993 | 203713_s_at | -0.7483 | 0.0000 | |
| GSE23400 | LLGL2 | 3993 | 203713_s_at | -0.4249 | 0.0000 | |
| GSE26886 | LLGL2 | 3993 | 1554006_a_at | -1.1485 | 0.0000 | |
| GSE29001 | LLGL2 | 3993 | 203713_s_at | -0.8209 | 0.0135 | |
| GSE38129 | LLGL2 | 3993 | 203713_s_at | -0.4476 | 0.0197 | |
| GSE45670 | LLGL2 | 3993 | 203713_s_at | -0.2066 | 0.3056 | |
| GSE53622 | LLGL2 | 3993 | 43749 | -0.9748 | 0.0000 | |
| GSE53624 | LLGL2 | 3993 | 43749 | -1.1961 | 0.0000 | |
| GSE63941 | LLGL2 | 3993 | 1554006_a_at | 1.4888 | 0.0098 | |
| GSE77861 | LLGL2 | 3993 | 203713_s_at | -0.2038 | 0.6062 | |
| GSE97050 | LLGL2 | 3993 | A_24_P307869 | 0.6856 | 0.2642 | |
| SRP007169 | LLGL2 | 3993 | RNAseq | -2.4585 | 0.0000 | |
| SRP008496 | LLGL2 | 3993 | RNAseq | -2.5329 | 0.0000 | |
| SRP064894 | LLGL2 | 3993 | RNAseq | -1.7362 | 0.0000 | |
| SRP133303 | LLGL2 | 3993 | RNAseq | -0.8107 | 0.0405 | |
| SRP159526 | LLGL2 | 3993 | RNAseq | -1.2087 | 0.0000 | |
| SRP193095 | LLGL2 | 3993 | RNAseq | -0.9281 | 0.0000 | |
| SRP219564 | LLGL2 | 3993 | RNAseq | -0.6519 | 0.4740 | |
| TCGA | LLGL2 | 3993 | RNAseq | -0.3309 | 0.0009 |
Upregulated datasets: 1; Downregulated datasets: 6.
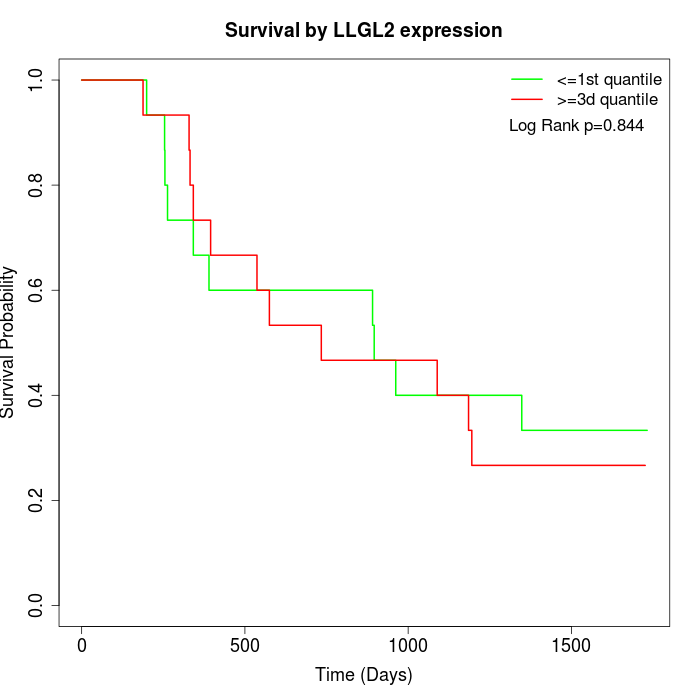
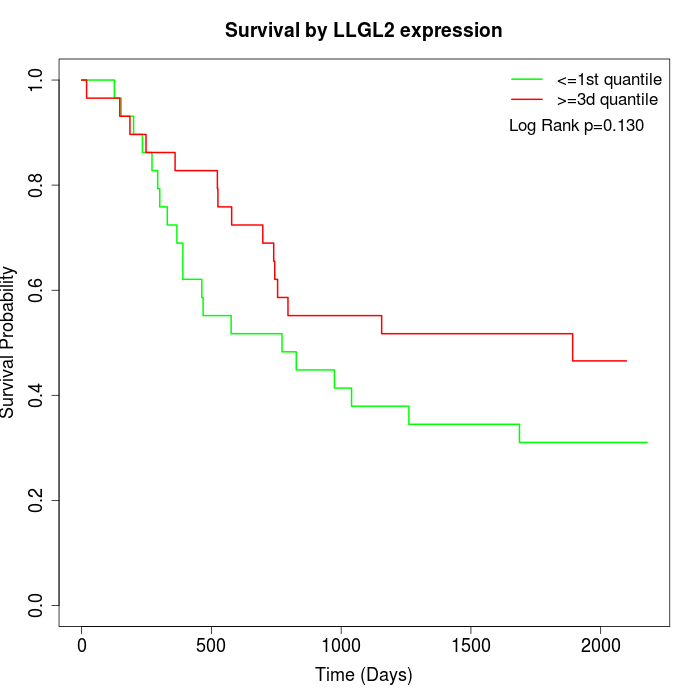
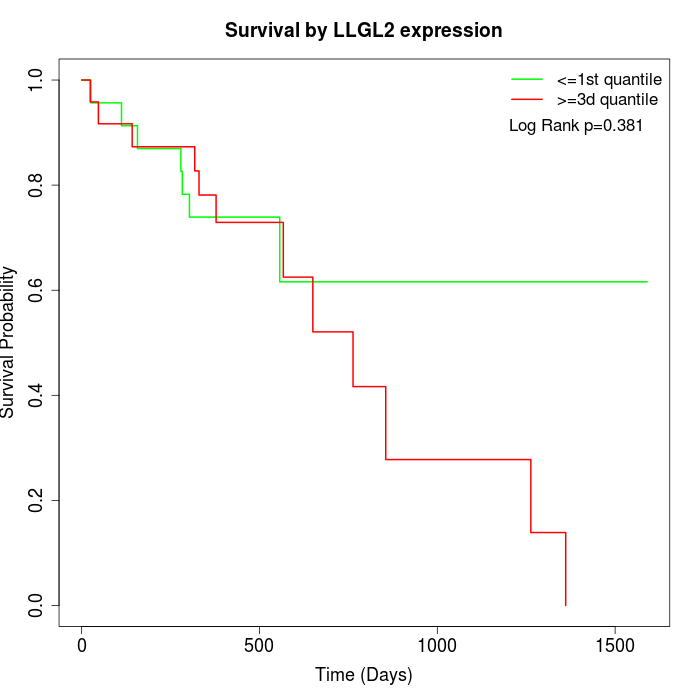
Survival by LLGL2 expression:
Note: Click image to view full size file.
Copy number change of LLGL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LLGL2 | 3993 | 6 | 2 | 22 | |
| GSE20123 | LLGL2 | 3993 | 6 | 2 | 22 | |
| GSE43470 | LLGL2 | 3993 | 5 | 3 | 35 | |
| GSE46452 | LLGL2 | 3993 | 34 | 0 | 25 | |
| GSE47630 | LLGL2 | 3993 | 8 | 1 | 31 | |
| GSE54993 | LLGL2 | 3993 | 2 | 5 | 63 | |
| GSE54994 | LLGL2 | 3993 | 10 | 4 | 39 | |
| GSE60625 | LLGL2 | 3993 | 6 | 0 | 5 | |
| GSE74703 | LLGL2 | 3993 | 5 | 1 | 30 | |
| GSE74704 | LLGL2 | 3993 | 4 | 1 | 15 | |
| TCGA | LLGL2 | 3993 | 32 | 9 | 55 |
Total number of gains: 118; Total number of losses: 28; Total Number of normals: 342.
Somatic mutations of LLGL2:
Generating mutation plots.
Highly correlated genes for LLGL2:
Showing top 20/1103 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LLGL2 | RECQL5 | 0.780324 | 11 | 0 | 11 |
| LLGL2 | EVPL | 0.769377 | 12 | 0 | 12 |
| LLGL2 | LIPH | 0.763479 | 8 | 0 | 8 |
| LLGL2 | MACC1 | 0.761907 | 7 | 0 | 7 |
| LLGL2 | EPHA1 | 0.752288 | 12 | 0 | 12 |
| LLGL2 | SMIM5 | 0.747798 | 7 | 0 | 7 |
| LLGL2 | SYTL1 | 0.746438 | 8 | 0 | 8 |
| LLGL2 | EPS8L1 | 0.746246 | 12 | 0 | 12 |
| LLGL2 | CLDN7 | 0.743293 | 12 | 0 | 12 |
| LLGL2 | NBEAL2 | 0.740321 | 12 | 0 | 12 |
| LLGL2 | ARHGAP27 | 0.737132 | 8 | 0 | 8 |
| LLGL2 | CAMSAP3 | 0.736292 | 8 | 0 | 8 |
| LLGL2 | ZDHHC13 | 0.734758 | 10 | 0 | 10 |
| LLGL2 | TST | 0.732146 | 9 | 0 | 9 |
| LLGL2 | NIPAL1 | 0.731675 | 3 | 0 | 3 |
| LLGL2 | LTA4H | 0.730658 | 3 | 0 | 3 |
| LLGL2 | ARAP2 | 0.728431 | 3 | 0 | 3 |
| LLGL2 | PLEKHM1 | 0.727711 | 10 | 0 | 10 |
| LLGL2 | SCNN1A | 0.726577 | 12 | 0 | 11 |
| LLGL2 | KLK11 | 0.726165 | 10 | 0 | 10 |
For details and further investigation, click here