| Full name: mitogen-activated protein kinase kinase kinase kinase 1 | Alias Symbol: HPK1 | ||
| Type: protein-coding gene | Cytoband: 19q13.1-q13.4 | ||
| Entrez ID: 11184 | HGNC ID: HGNC:6863 | Ensembl Gene: ENSG00000104814 | OMIM ID: 601983 |
| Drug and gene relationship at DGIdb | |||
MAP4K1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway |
Expression of MAP4K1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP4K1 | 11184 | 214339_s_at | 0.2495 | 0.7760 | |
| GSE20347 | MAP4K1 | 11184 | 214339_s_at | 0.2073 | 0.1010 | |
| GSE23400 | MAP4K1 | 11184 | 206296_x_at | -0.0135 | 0.8034 | |
| GSE26886 | MAP4K1 | 11184 | 214339_s_at | 0.2710 | 0.2266 | |
| GSE29001 | MAP4K1 | 11184 | 214339_s_at | 0.2143 | 0.4644 | |
| GSE38129 | MAP4K1 | 11184 | 206296_x_at | 0.1298 | 0.5196 | |
| GSE45670 | MAP4K1 | 11184 | 214339_s_at | 0.1037 | 0.4140 | |
| GSE53622 | MAP4K1 | 11184 | 62308 | -0.0767 | 0.4186 | |
| GSE53624 | MAP4K1 | 11184 | 62308 | -0.0794 | 0.3210 | |
| GSE63941 | MAP4K1 | 11184 | 214339_s_at | 1.4267 | 0.0830 | |
| GSE77861 | MAP4K1 | 11184 | 214339_s_at | -0.0075 | 0.9667 | |
| GSE97050 | MAP4K1 | 11184 | A_23_P5002 | 0.5260 | 0.1790 | |
| SRP007169 | MAP4K1 | 11184 | RNAseq | 0.9475 | 0.1884 | |
| SRP064894 | MAP4K1 | 11184 | RNAseq | 0.1740 | 0.5639 | |
| SRP133303 | MAP4K1 | 11184 | RNAseq | -0.5459 | 0.0792 | |
| SRP159526 | MAP4K1 | 11184 | RNAseq | -0.9606 | 0.1572 | |
| SRP193095 | MAP4K1 | 11184 | RNAseq | -0.4445 | 0.0380 | |
| SRP219564 | MAP4K1 | 11184 | RNAseq | 0.2021 | 0.5861 | |
| TCGA | MAP4K1 | 11184 | RNAseq | 0.1166 | 0.6084 |
Upregulated datasets: 0; Downregulated datasets: 0.
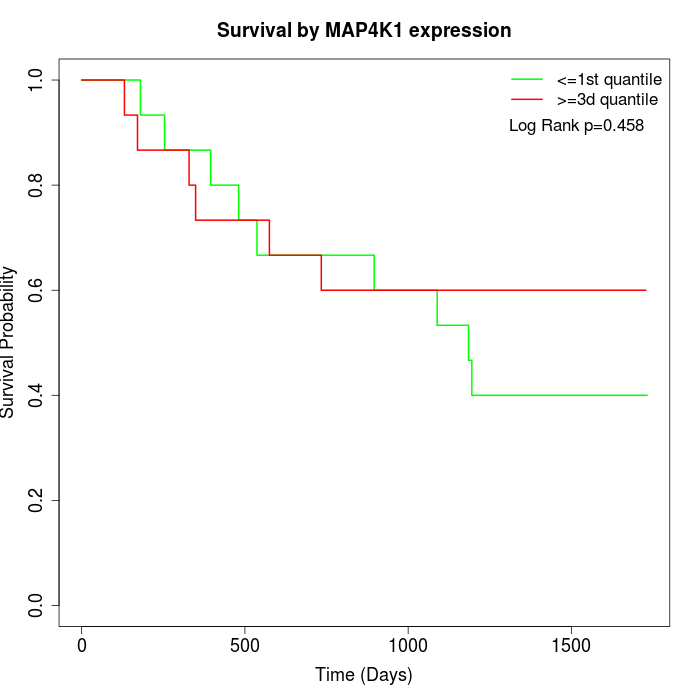
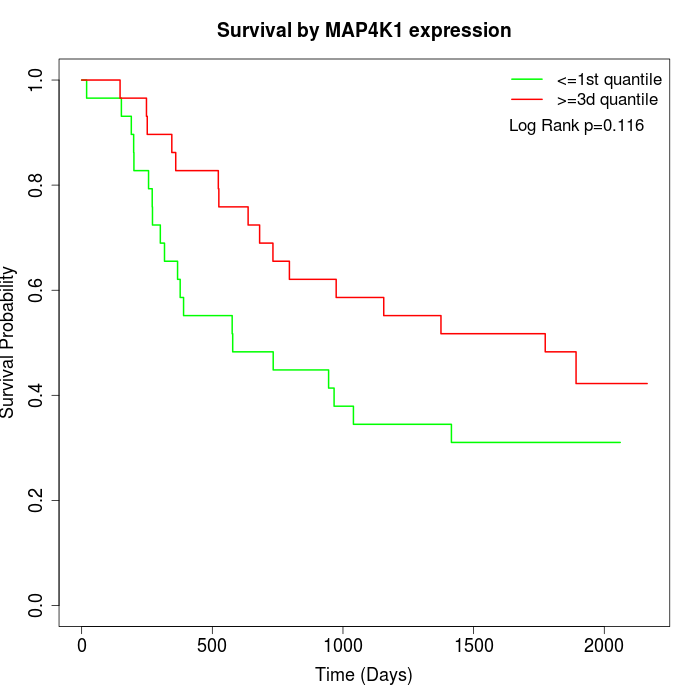
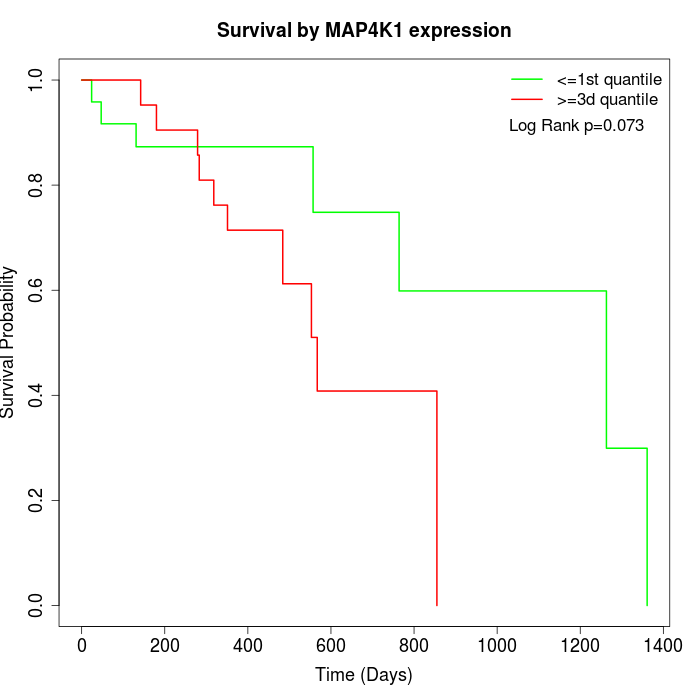
Survival by MAP4K1 expression:
Note: Click image to view full size file.
Copy number change of MAP4K1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP4K1 | 11184 | 7 | 4 | 19 | |
| GSE20123 | MAP4K1 | 11184 | 7 | 3 | 20 | |
| GSE43470 | MAP4K1 | 11184 | 4 | 10 | 29 | |
| GSE46452 | MAP4K1 | 11184 | 47 | 2 | 10 | |
| GSE47630 | MAP4K1 | 11184 | 9 | 5 | 26 | |
| GSE54993 | MAP4K1 | 11184 | 17 | 3 | 50 | |
| GSE54994 | MAP4K1 | 11184 | 6 | 9 | 38 | |
| GSE60625 | MAP4K1 | 11184 | 9 | 0 | 2 | |
| GSE74703 | MAP4K1 | 11184 | 4 | 6 | 26 | |
| GSE74704 | MAP4K1 | 11184 | 7 | 1 | 12 | |
| TCGA | MAP4K1 | 11184 | 16 | 15 | 65 |
Total number of gains: 133; Total number of losses: 58; Total Number of normals: 297.
Somatic mutations of MAP4K1:
Generating mutation plots.
Highly correlated genes for MAP4K1:
Showing top 20/223 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP4K1 | TBC1D10C | 0.726152 | 5 | 0 | 5 |
| MAP4K1 | PARVG | 0.711064 | 4 | 0 | 4 |
| MAP4K1 | LYPD1 | 0.703345 | 3 | 0 | 3 |
| MAP4K1 | MCOLN2 | 0.702481 | 4 | 0 | 3 |
| MAP4K1 | TAPBPL | 0.680017 | 3 | 0 | 3 |
| MAP4K1 | ABCC5 | 0.667892 | 3 | 0 | 3 |
| MAP4K1 | HHAT | 0.66546 | 3 | 0 | 3 |
| MAP4K1 | PLXNA3 | 0.660995 | 3 | 0 | 3 |
| MAP4K1 | FMNL1 | 0.655405 | 6 | 0 | 4 |
| MAP4K1 | KCNA3 | 0.651929 | 3 | 0 | 3 |
| MAP4K1 | CPNE5 | 0.650316 | 5 | 0 | 5 |
| MAP4K1 | PGPEP1 | 0.650185 | 3 | 0 | 3 |
| MAP4K1 | SNX20 | 0.650018 | 3 | 0 | 3 |
| MAP4K1 | ITGAL | 0.645119 | 9 | 0 | 7 |
| MAP4K1 | LAX1 | 0.643568 | 5 | 0 | 4 |
| MAP4K1 | EPHB2 | 0.640382 | 4 | 0 | 3 |
| MAP4K1 | KIR2DL5A | 0.638693 | 3 | 0 | 3 |
| MAP4K1 | HSH2D | 0.638448 | 5 | 0 | 4 |
| MAP4K1 | YIF1B | 0.63718 | 4 | 0 | 3 |
| MAP4K1 | DTNB | 0.63657 | 4 | 0 | 4 |
For details and further investigation, click here