| Full name: hedgehog acyltransferase | Alias Symbol: FLJ10724|MART-2|MART2|Skn|ski|rasp|sit|GUP2 | ||
| Type: protein-coding gene | Cytoband: 1q32.2 | ||
| Entrez ID: 55733 | HGNC ID: HGNC:18270 | Ensembl Gene: ENSG00000054392 | OMIM ID: 605743 |
| Drug and gene relationship at DGIdb | |||
Expression of HHAT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HHAT | 55733 | 219687_at | 0.2784 | 0.5452 | |
| GSE20347 | HHAT | 55733 | 219687_at | 0.2372 | 0.1027 | |
| GSE23400 | HHAT | 55733 | 219687_at | 0.0623 | 0.2037 | |
| GSE26886 | HHAT | 55733 | 219687_at | 0.4488 | 0.1315 | |
| GSE29001 | HHAT | 55733 | 219687_at | 0.3177 | 0.3796 | |
| GSE38129 | HHAT | 55733 | 219687_at | 0.3883 | 0.0084 | |
| GSE45670 | HHAT | 55733 | 219687_at | 0.1898 | 0.4283 | |
| GSE53622 | HHAT | 55733 | 163226 | -0.0455 | 0.7168 | |
| GSE53624 | HHAT | 55733 | 163226 | 0.0358 | 0.7189 | |
| GSE63941 | HHAT | 55733 | 219687_at | -0.9049 | 0.0903 | |
| GSE77861 | HHAT | 55733 | 219687_at | 0.0655 | 0.6934 | |
| GSE97050 | HHAT | 55733 | A_23_P136355 | 0.2930 | 0.2401 | |
| SRP007169 | HHAT | 55733 | RNAseq | 0.6156 | 0.2947 | |
| SRP008496 | HHAT | 55733 | RNAseq | 0.6240 | 0.1246 | |
| SRP064894 | HHAT | 55733 | RNAseq | 0.3926 | 0.0737 | |
| SRP133303 | HHAT | 55733 | RNAseq | -0.2588 | 0.2802 | |
| SRP159526 | HHAT | 55733 | RNAseq | 0.2648 | 0.5644 | |
| SRP193095 | HHAT | 55733 | RNAseq | -0.1387 | 0.4520 | |
| SRP219564 | HHAT | 55733 | RNAseq | 0.1216 | 0.7681 | |
| TCGA | HHAT | 55733 | RNAseq | 0.0792 | 0.5249 |
Upregulated datasets: 0; Downregulated datasets: 0.
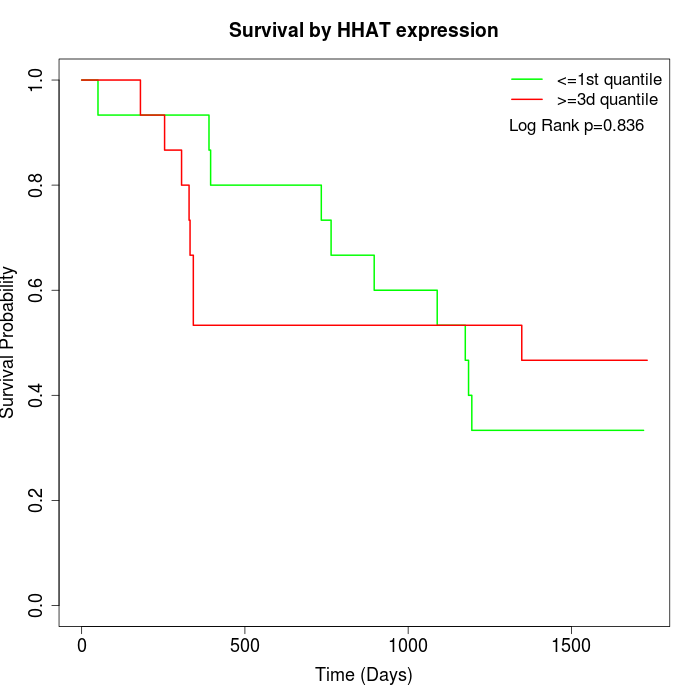
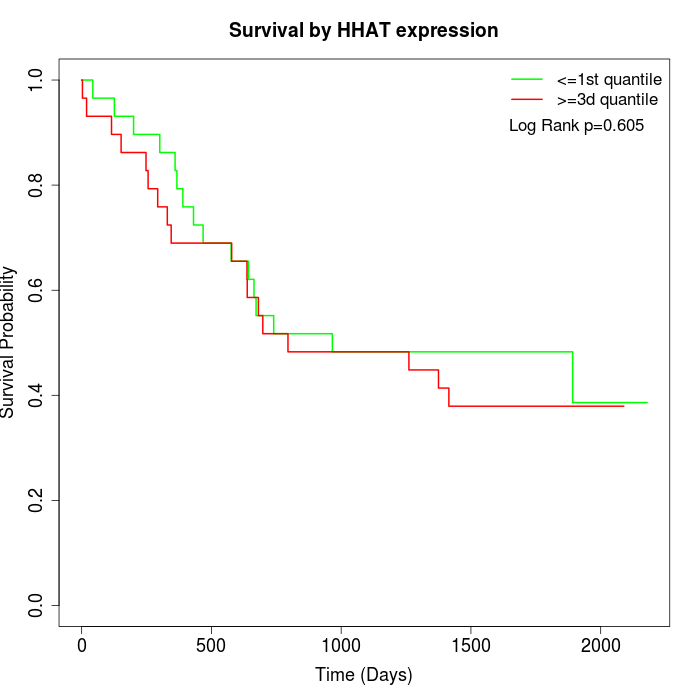
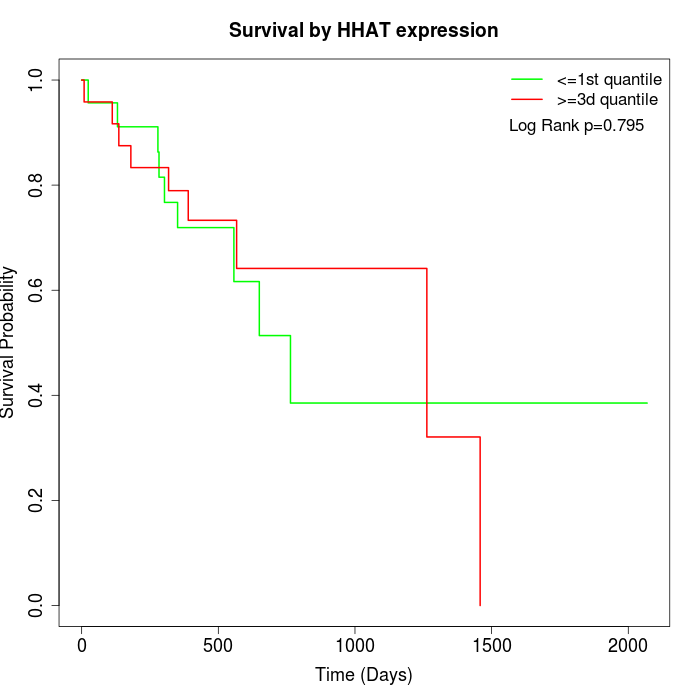
Survival by HHAT expression:
Note: Click image to view full size file.
Copy number change of HHAT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HHAT | 55733 | 11 | 0 | 19 | |
| GSE20123 | HHAT | 55733 | 10 | 0 | 20 | |
| GSE43470 | HHAT | 55733 | 7 | 0 | 36 | |
| GSE46452 | HHAT | 55733 | 3 | 1 | 55 | |
| GSE47630 | HHAT | 55733 | 15 | 0 | 25 | |
| GSE54993 | HHAT | 55733 | 0 | 6 | 64 | |
| GSE54994 | HHAT | 55733 | 14 | 0 | 39 | |
| GSE60625 | HHAT | 55733 | 0 | 0 | 11 | |
| GSE74703 | HHAT | 55733 | 7 | 0 | 29 | |
| GSE74704 | HHAT | 55733 | 4 | 0 | 16 | |
| TCGA | HHAT | 55733 | 43 | 5 | 48 |
Total number of gains: 114; Total number of losses: 12; Total Number of normals: 362.
Somatic mutations of HHAT:
Generating mutation plots.
Highly correlated genes for HHAT:
Showing top 20/242 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HHAT | FBXO27 | 0.795261 | 3 | 0 | 3 |
| HHAT | KIF3B | 0.733527 | 3 | 0 | 3 |
| HHAT | FGF11 | 0.719195 | 3 | 0 | 3 |
| HHAT | HOXD8 | 0.712714 | 3 | 0 | 3 |
| HHAT | QSOX2 | 0.7058 | 3 | 0 | 3 |
| HHAT | ZNF146 | 0.702772 | 4 | 0 | 3 |
| HHAT | TRMT2A | 0.699057 | 3 | 0 | 3 |
| HHAT | CYHR1 | 0.694537 | 4 | 0 | 3 |
| HHAT | ANKRD23 | 0.691026 | 3 | 0 | 3 |
| HHAT | MRPS9 | 0.685695 | 3 | 0 | 3 |
| HHAT | DGKD | 0.675955 | 3 | 0 | 3 |
| HHAT | YWHAE | 0.673473 | 4 | 0 | 3 |
| HHAT | SP3 | 0.672091 | 3 | 0 | 3 |
| HHAT | HMGXB4 | 0.671578 | 3 | 0 | 3 |
| HHAT | RAC3 | 0.66935 | 3 | 0 | 3 |
| HHAT | SLC10A3 | 0.667917 | 5 | 0 | 3 |
| HHAT | MAP4K1 | 0.66546 | 3 | 0 | 3 |
| HHAT | TMEM132A | 0.662845 | 3 | 0 | 3 |
| HHAT | CHD7 | 0.657718 | 4 | 0 | 3 |
| HHAT | ZNF473 | 0.652769 | 3 | 0 | 3 |
For details and further investigation, click here