| Full name: MYC associated factor X | Alias Symbol: bHLHd4|bHLHd5|bHLHd6|bHLHd7|bHLHd8 | ||
| Type: protein-coding gene | Cytoband: 14q23.3 | ||
| Entrez ID: 4149 | HGNC ID: HGNC:6913 | Ensembl Gene: ENSG00000125952 | OMIM ID: 154950 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
MAX involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa05200 | Pathways in cancer | |
| hsa05222 | Small cell lung cancer |
Expression of MAX:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAX | 4149 | 209332_s_at | -0.3731 | 0.1673 | |
| GSE20347 | MAX | 4149 | 209332_s_at | -0.2933 | 0.0353 | |
| GSE23400 | MAX | 4149 | 209332_s_at | -0.0980 | 0.1786 | |
| GSE26886 | MAX | 4149 | 209332_s_at | -0.8363 | 0.0000 | |
| GSE29001 | MAX | 4149 | 209332_s_at | 0.0645 | 0.7830 | |
| GSE38129 | MAX | 4149 | 209332_s_at | -0.2981 | 0.0063 | |
| GSE45670 | MAX | 4149 | 209332_s_at | -0.3365 | 0.0087 | |
| GSE53622 | MAX | 4149 | 28909 | -0.2453 | 0.0001 | |
| GSE53624 | MAX | 4149 | 28909 | -0.3190 | 0.0000 | |
| GSE63941 | MAX | 4149 | 209332_s_at | -0.2656 | 0.5650 | |
| GSE77861 | MAX | 4149 | 209332_s_at | -0.3350 | 0.2364 | |
| GSE97050 | MAX | 4149 | A_23_P151662 | 0.0968 | 0.8089 | |
| SRP007169 | MAX | 4149 | RNAseq | -1.2888 | 0.0004 | |
| SRP008496 | MAX | 4149 | RNAseq | -1.2813 | 0.0000 | |
| SRP064894 | MAX | 4149 | RNAseq | -0.6510 | 0.0000 | |
| SRP133303 | MAX | 4149 | RNAseq | -0.5457 | 0.0015 | |
| SRP159526 | MAX | 4149 | RNAseq | -0.7035 | 0.0005 | |
| SRP193095 | MAX | 4149 | RNAseq | -0.5340 | 0.0000 | |
| SRP219564 | MAX | 4149 | RNAseq | -0.4419 | 0.1558 | |
| TCGA | MAX | 4149 | RNAseq | 0.0617 | 0.2183 |
Upregulated datasets: 0; Downregulated datasets: 2.
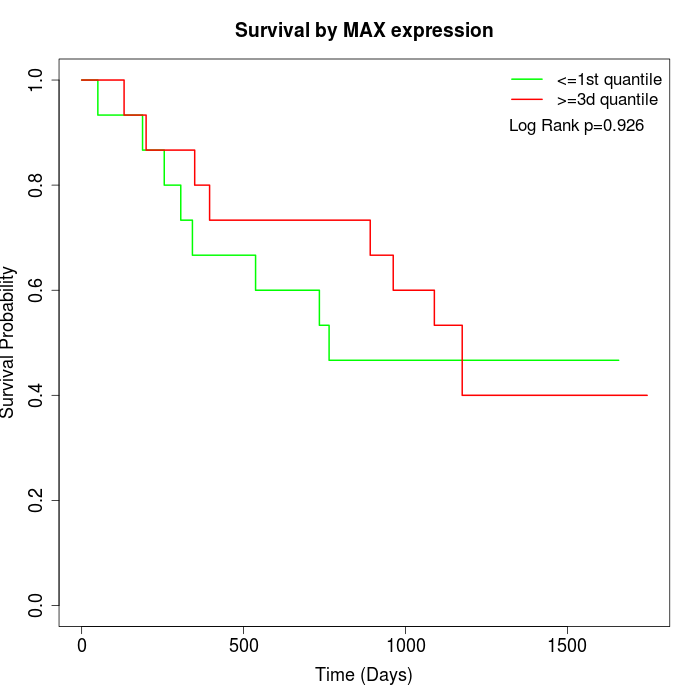
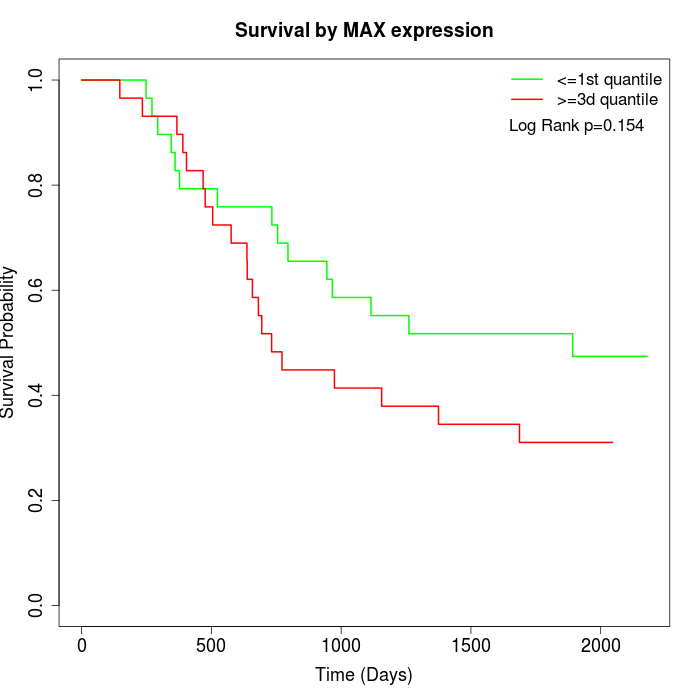
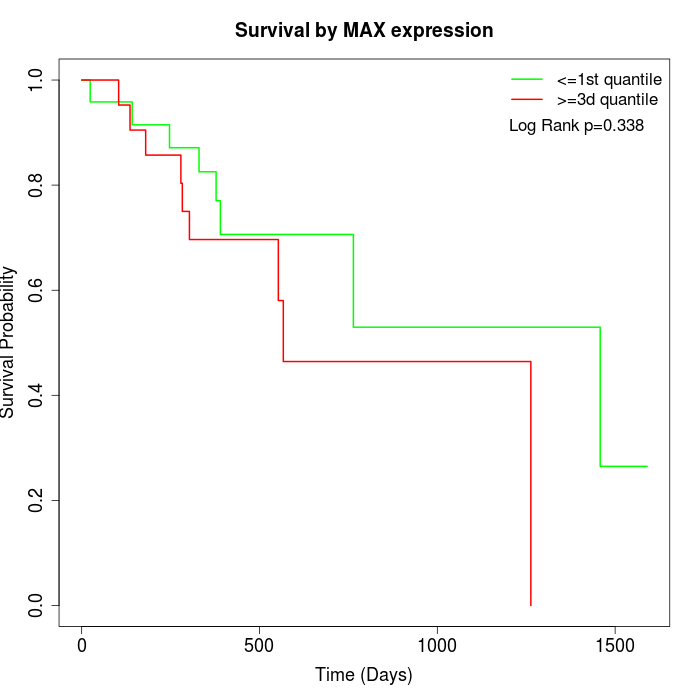
Survival by MAX expression:
Note: Click image to view full size file.
Copy number change of MAX:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAX | 4149 | 7 | 3 | 20 | |
| GSE20123 | MAX | 4149 | 7 | 3 | 20 | |
| GSE43470 | MAX | 4149 | 8 | 2 | 33 | |
| GSE46452 | MAX | 4149 | 16 | 3 | 40 | |
| GSE47630 | MAX | 4149 | 11 | 9 | 20 | |
| GSE54993 | MAX | 4149 | 3 | 9 | 58 | |
| GSE54994 | MAX | 4149 | 18 | 5 | 30 | |
| GSE60625 | MAX | 4149 | 0 | 2 | 9 | |
| GSE74703 | MAX | 4149 | 7 | 2 | 27 | |
| GSE74704 | MAX | 4149 | 3 | 2 | 15 | |
| TCGA | MAX | 4149 | 32 | 14 | 50 |
Total number of gains: 112; Total number of losses: 54; Total Number of normals: 322.
Somatic mutations of MAX:
Generating mutation plots.
Highly correlated genes for MAX:
Showing top 20/153 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAX | TMEM220 | 0.70565 | 3 | 0 | 3 |
| MAX | KCTD15 | 0.653947 | 3 | 0 | 3 |
| MAX | BLOC1S2 | 0.652768 | 3 | 0 | 3 |
| MAX | NIPAL2 | 0.646685 | 3 | 0 | 3 |
| MAX | STX19 | 0.631937 | 3 | 0 | 3 |
| MAX | SLC7A1 | 0.629414 | 3 | 0 | 3 |
| MAX | CDC42BPB | 0.616299 | 4 | 0 | 4 |
| MAX | KLK13 | 0.615971 | 4 | 0 | 3 |
| MAX | ZFYVE1 | 0.614776 | 6 | 0 | 5 |
| MAX | RNF32 | 0.611067 | 3 | 0 | 3 |
| MAX | PIK3C2G | 0.606845 | 5 | 0 | 3 |
| MAX | ZNF407 | 0.604075 | 3 | 0 | 3 |
| MAX | CST11 | 0.600594 | 3 | 0 | 3 |
| MAX | FUT11 | 0.59775 | 3 | 0 | 3 |
| MAX | HDHD2 | 0.595781 | 3 | 0 | 3 |
| MAX | DHRS12 | 0.594007 | 4 | 0 | 3 |
| MAX | CLIC3 | 0.592452 | 4 | 0 | 3 |
| MAX | ATP6V1D | 0.591589 | 6 | 0 | 4 |
| MAX | ISM1 | 0.588646 | 3 | 0 | 3 |
| MAX | C14orf28 | 0.584674 | 4 | 0 | 3 |
For details and further investigation, click here