| Full name: mitochondrial fission factor | Alias Symbol: GL004 | ||
| Type: protein-coding gene | Cytoband: 2q36.3 | ||
| Entrez ID: 56947 | HGNC ID: HGNC:24858 | Ensembl Gene: ENSG00000168958 | OMIM ID: 614785 |
| Drug and gene relationship at DGIdb | |||
Expression of MFF:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MFF | 56947 | 222832_s_at | -0.2540 | 0.5174 | |
| GSE20347 | MFF | 56947 | 219137_s_at | -0.1232 | 0.5282 | |
| GSE23400 | MFF | 56947 | 219137_s_at | 0.0292 | 0.7798 | |
| GSE26886 | MFF | 56947 | 222832_s_at | 0.6078 | 0.0009 | |
| GSE29001 | MFF | 56947 | 219137_s_at | 0.2004 | 0.6070 | |
| GSE38129 | MFF | 56947 | 219137_s_at | -0.1999 | 0.2420 | |
| GSE45670 | MFF | 56947 | 222832_s_at | 0.0489 | 0.7566 | |
| GSE53622 | MFF | 56947 | 29220 | 0.0640 | 0.3218 | |
| GSE53624 | MFF | 56947 | 29220 | 0.1352 | 0.0028 | |
| GSE63941 | MFF | 56947 | 222832_s_at | -0.0141 | 0.9755 | |
| GSE77861 | MFF | 56947 | 222832_s_at | 0.2147 | 0.5344 | |
| GSE97050 | MFF | 56947 | A_23_P405148 | -0.0988 | 0.7732 | |
| SRP007169 | MFF | 56947 | RNAseq | -0.3121 | 0.4800 | |
| SRP008496 | MFF | 56947 | RNAseq | -0.0529 | 0.8453 | |
| SRP064894 | MFF | 56947 | RNAseq | -0.0109 | 0.9380 | |
| SRP133303 | MFF | 56947 | RNAseq | 0.1425 | 0.1930 | |
| SRP159526 | MFF | 56947 | RNAseq | 0.1673 | 0.3581 | |
| SRP193095 | MFF | 56947 | RNAseq | -0.0411 | 0.7231 | |
| SRP219564 | MFF | 56947 | RNAseq | -0.3389 | 0.3358 | |
| TCGA | MFF | 56947 | RNAseq | 0.0095 | 0.8606 |
Upregulated datasets: 0; Downregulated datasets: 0.
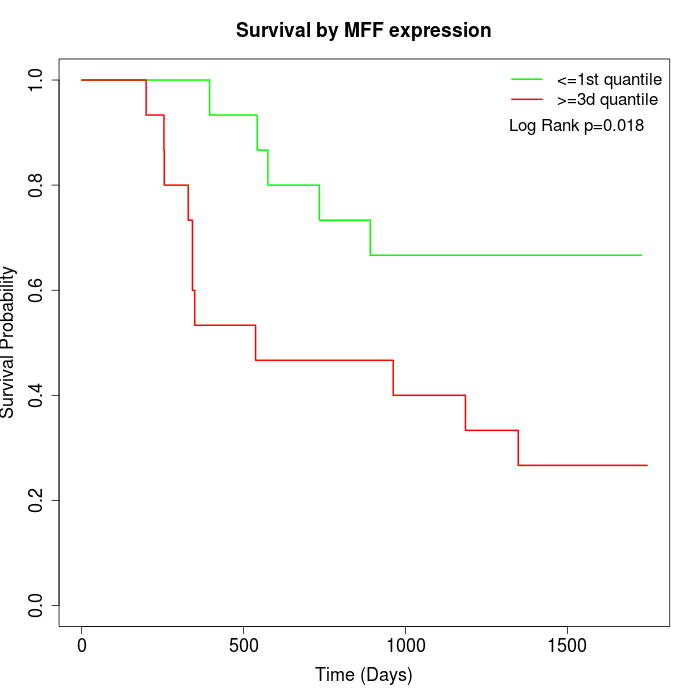
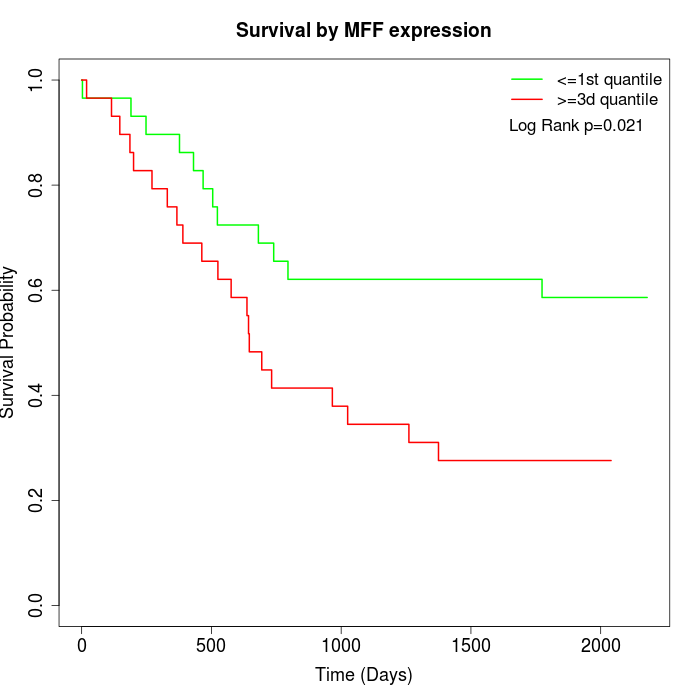
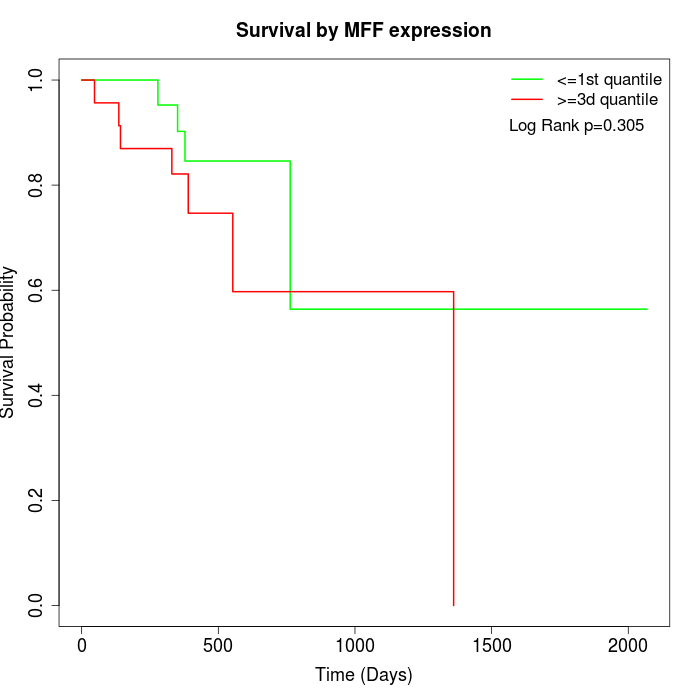
Survival by MFF expression:
Note: Click image to view full size file.
Copy number change of MFF:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MFF | 56947 | 2 | 12 | 16 | |
| GSE20123 | MFF | 56947 | 2 | 12 | 16 | |
| GSE43470 | MFF | 56947 | 0 | 7 | 36 | |
| GSE46452 | MFF | 56947 | 0 | 5 | 54 | |
| GSE47630 | MFF | 56947 | 4 | 5 | 31 | |
| GSE54993 | MFF | 56947 | 1 | 2 | 67 | |
| GSE54994 | MFF | 56947 | 6 | 11 | 36 | |
| GSE60625 | MFF | 56947 | 0 | 3 | 8 | |
| GSE74703 | MFF | 56947 | 0 | 6 | 30 | |
| GSE74704 | MFF | 56947 | 2 | 6 | 12 | |
| TCGA | MFF | 56947 | 13 | 26 | 57 |
Total number of gains: 30; Total number of losses: 95; Total Number of normals: 363.
Somatic mutations of MFF:
Generating mutation plots.
Highly correlated genes for MFF:
Showing top 20/370 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MFF | ALDOA | 0.791974 | 3 | 0 | 3 |
| MFF | NANOS1 | 0.759738 | 3 | 0 | 3 |
| MFF | SMARCA5 | 0.751554 | 4 | 0 | 4 |
| MFF | HERPUD2 | 0.751206 | 3 | 0 | 3 |
| MFF | PRKACB | 0.73046 | 4 | 0 | 4 |
| MFF | SCAPER | 0.719975 | 3 | 0 | 3 |
| MFF | TOR1B | 0.717038 | 3 | 0 | 3 |
| MFF | LMO1 | 0.716242 | 3 | 0 | 3 |
| MFF | FRY | 0.713994 | 3 | 0 | 3 |
| MFF | ZNF558 | 0.708939 | 3 | 0 | 3 |
| MFF | POP5 | 0.707958 | 3 | 0 | 3 |
| MFF | PCGF2 | 0.707236 | 3 | 0 | 3 |
| MFF | SLC25A46 | 0.706455 | 3 | 0 | 3 |
| MFF | CDO1 | 0.70579 | 3 | 0 | 3 |
| MFF | POLR2G | 0.70415 | 3 | 0 | 3 |
| MFF | ADAT1 | 0.698566 | 3 | 0 | 3 |
| MFF | TOM1L1 | 0.697653 | 4 | 0 | 4 |
| MFF | CDKL3 | 0.692994 | 3 | 0 | 3 |
| MFF | GADD45A | 0.692597 | 3 | 0 | 3 |
| MFF | DIXDC1 | 0.691264 | 3 | 0 | 3 |
For details and further investigation, click here