| Full name: mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase | Alias Symbol: GNT-1|GLCNAC-TI|GnTI | ||
| Type: protein-coding gene | Cytoband: 5q35.3 | ||
| Entrez ID: 4245 | HGNC ID: HGNC:7044 | Ensembl Gene: ENSG00000131446 | OMIM ID: 160995 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of MGAT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MGAT1 | 4245 | 201126_s_at | -0.0124 | 0.9786 | |
| GSE20347 | MGAT1 | 4245 | 201126_s_at | -0.5751 | 0.0000 | |
| GSE23400 | MGAT1 | 4245 | 201126_s_at | -0.3902 | 0.0000 | |
| GSE26886 | MGAT1 | 4245 | 201126_s_at | -0.9456 | 0.0007 | |
| GSE29001 | MGAT1 | 4245 | 201126_s_at | -0.7166 | 0.0026 | |
| GSE38129 | MGAT1 | 4245 | 201126_s_at | -0.4770 | 0.0000 | |
| GSE45670 | MGAT1 | 4245 | 201126_s_at | -0.4150 | 0.0062 | |
| GSE53622 | MGAT1 | 4245 | 9374 | -0.4595 | 0.0000 | |
| GSE53624 | MGAT1 | 4245 | 9374 | -0.4820 | 0.0000 | |
| GSE63941 | MGAT1 | 4245 | 201126_s_at | -1.0180 | 0.0236 | |
| GSE77861 | MGAT1 | 4245 | 201126_s_at | -0.2477 | 0.2970 | |
| GSE97050 | MGAT1 | 4245 | A_33_P3317664 | -0.3638 | 0.3889 | |
| SRP007169 | MGAT1 | 4245 | RNAseq | -2.0595 | 0.0000 | |
| SRP008496 | MGAT1 | 4245 | RNAseq | -1.9652 | 0.0000 | |
| SRP064894 | MGAT1 | 4245 | RNAseq | -0.2214 | 0.3516 | |
| SRP133303 | MGAT1 | 4245 | RNAseq | -0.2154 | 0.2564 | |
| SRP159526 | MGAT1 | 4245 | RNAseq | -0.6478 | 0.0861 | |
| SRP193095 | MGAT1 | 4245 | RNAseq | -0.7472 | 0.0000 | |
| SRP219564 | MGAT1 | 4245 | RNAseq | -0.2641 | 0.4150 | |
| TCGA | MGAT1 | 4245 | RNAseq | -0.0515 | 0.2324 |
Upregulated datasets: 0; Downregulated datasets: 3.
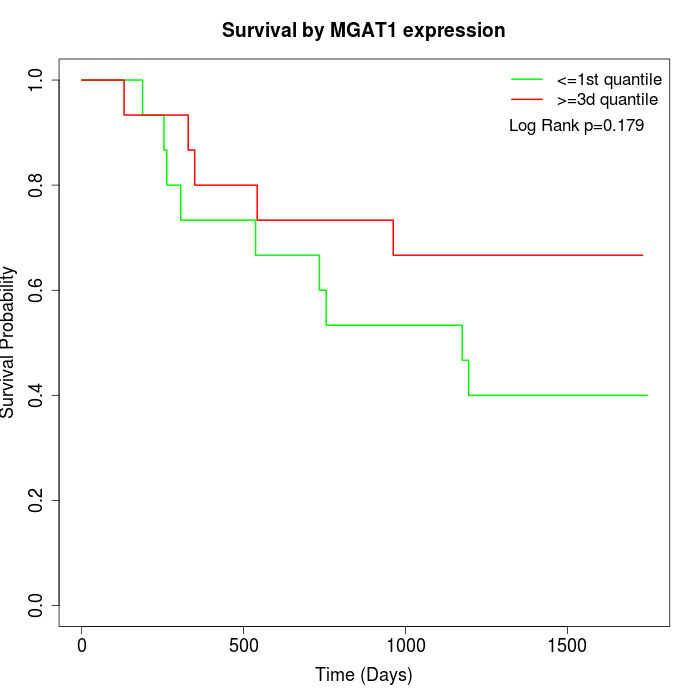
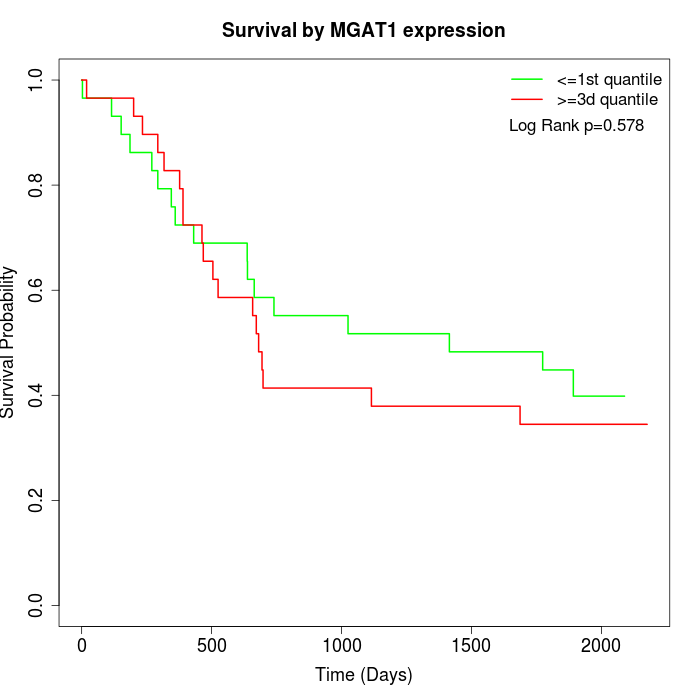
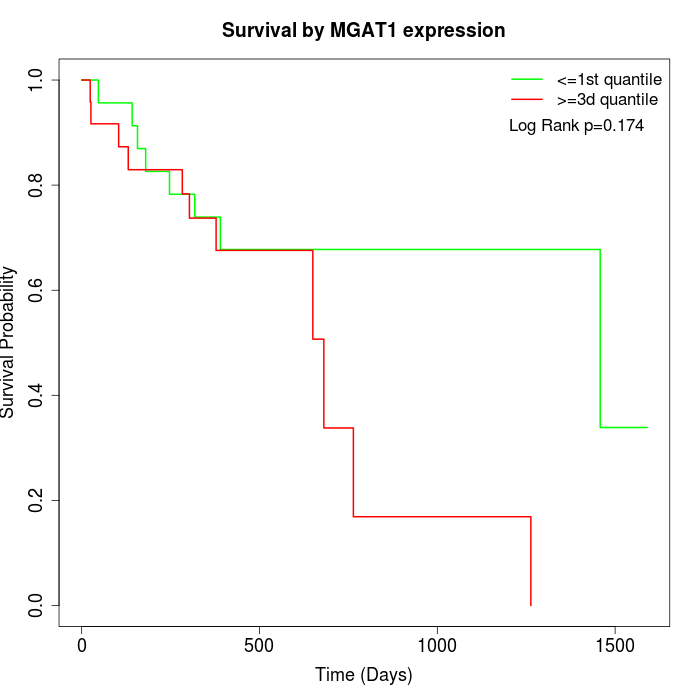
Survival by MGAT1 expression:
Note: Click image to view full size file.
Copy number change of MGAT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MGAT1 | 4245 | 2 | 13 | 15 | |
| GSE20123 | MGAT1 | 4245 | 2 | 13 | 15 | |
| GSE43470 | MGAT1 | 4245 | 1 | 10 | 32 | |
| GSE46452 | MGAT1 | 4245 | 0 | 27 | 32 | |
| GSE47630 | MGAT1 | 4245 | 1 | 20 | 19 | |
| GSE54993 | MGAT1 | 4245 | 10 | 2 | 58 | |
| GSE54994 | MGAT1 | 4245 | 3 | 18 | 32 | |
| GSE60625 | MGAT1 | 4245 | 1 | 0 | 10 | |
| GSE74703 | MGAT1 | 4245 | 1 | 6 | 29 | |
| GSE74704 | MGAT1 | 4245 | 1 | 6 | 13 | |
| TCGA | MGAT1 | 4245 | 12 | 34 | 50 |
Total number of gains: 34; Total number of losses: 149; Total Number of normals: 305.
Somatic mutations of MGAT1:
Generating mutation plots.
Highly correlated genes for MGAT1:
Showing top 20/336 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MGAT1 | FAM171A2 | 0.684796 | 3 | 0 | 3 |
| MGAT1 | B3GNT8 | 0.683008 | 4 | 0 | 4 |
| MGAT1 | VAT1 | 0.676544 | 10 | 0 | 10 |
| MGAT1 | ZNRF1 | 0.669672 | 4 | 0 | 4 |
| MGAT1 | ABLIM3 | 0.669331 | 11 | 0 | 10 |
| MGAT1 | SEMA3B | 0.663374 | 4 | 0 | 4 |
| MGAT1 | DPYSL4 | 0.663021 | 3 | 0 | 3 |
| MGAT1 | PMM1 | 0.658031 | 12 | 0 | 11 |
| MGAT1 | SERPINB6 | 0.653289 | 9 | 0 | 8 |
| MGAT1 | CHI3L1 | 0.64862 | 3 | 0 | 3 |
| MGAT1 | ZNF524 | 0.647938 | 4 | 0 | 4 |
| MGAT1 | SEC24A | 0.645107 | 5 | 0 | 4 |
| MGAT1 | ETHE1 | 0.64293 | 12 | 0 | 10 |
| MGAT1 | PHACTR4 | 0.641695 | 10 | 0 | 9 |
| MGAT1 | ESAM | 0.638492 | 6 | 0 | 5 |
| MGAT1 | SLC35C1 | 0.636588 | 10 | 0 | 8 |
| MGAT1 | CNN3 | 0.634961 | 9 | 0 | 8 |
| MGAT1 | CRISP3 | 0.633936 | 8 | 0 | 7 |
| MGAT1 | SH3GL1 | 0.631364 | 10 | 0 | 9 |
| MGAT1 | CDC42BPB | 0.629495 | 3 | 0 | 3 |
For details and further investigation, click here