| Full name: modulator of VRAC current 1 | Alias Symbol: MLC|KIAA0027|LVM|VL | ||
| Type: protein-coding gene | Cytoband: 22q13.33 | ||
| Entrez ID: 23209 | HGNC ID: HGNC:17082 | Ensembl Gene: ENSG00000100427 | OMIM ID: 605908 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of MLC1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MLC1 | 23209 | 213395_at | -0.0062 | 0.9879 | |
| GSE20347 | MLC1 | 23209 | 213395_at | 0.0265 | 0.7825 | |
| GSE23400 | MLC1 | 23209 | 213395_at | -0.0341 | 0.4231 | |
| GSE26886 | MLC1 | 23209 | 213395_at | 0.2826 | 0.0201 | |
| GSE29001 | MLC1 | 23209 | 213395_at | -0.1286 | 0.4655 | |
| GSE38129 | MLC1 | 23209 | 213395_at | 0.0818 | 0.2602 | |
| GSE45670 | MLC1 | 23209 | 213395_at | 0.1268 | 0.3284 | |
| GSE63941 | MLC1 | 23209 | 213395_at | -0.1927 | 0.4044 | |
| GSE77861 | MLC1 | 23209 | 213395_at | 0.0640 | 0.6802 | |
| TCGA | MLC1 | 23209 | RNAseq | 1.1968 | 0.0095 |
Upregulated datasets: 1; Downregulated datasets: 0.
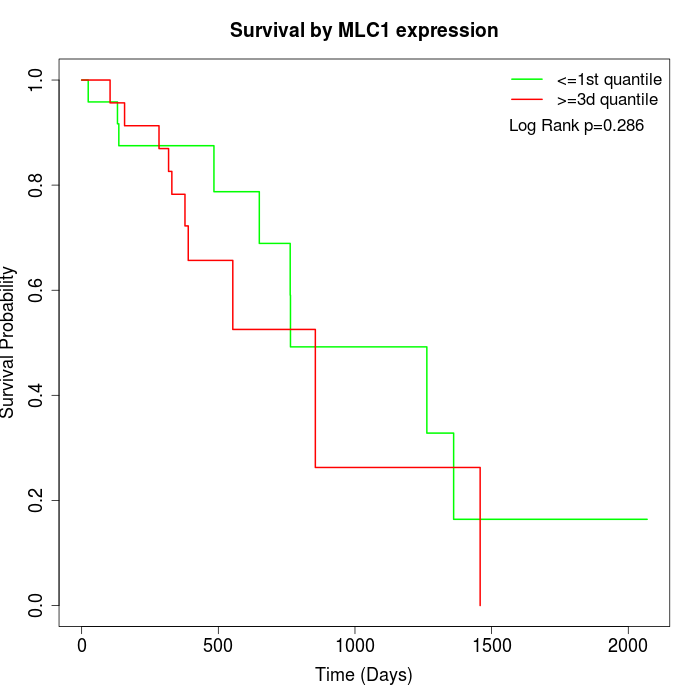
Survival by MLC1 expression:
Note: Click image to view full size file.
Copy number change of MLC1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MLC1 | 23209 | 3 | 8 | 19 | |
| GSE20123 | MLC1 | 23209 | 3 | 8 | 19 | |
| GSE43470 | MLC1 | 23209 | 2 | 8 | 33 | |
| GSE46452 | MLC1 | 23209 | 31 | 2 | 26 | |
| GSE47630 | MLC1 | 23209 | 9 | 4 | 27 | |
| GSE54993 | MLC1 | 23209 | 4 | 6 | 60 | |
| GSE54994 | MLC1 | 23209 | 11 | 8 | 34 | |
| GSE60625 | MLC1 | 23209 | 5 | 0 | 6 | |
| GSE74703 | MLC1 | 23209 | 2 | 6 | 28 | |
| GSE74704 | MLC1 | 23209 | 1 | 4 | 15 | |
| TCGA | MLC1 | 23209 | 26 | 16 | 54 |
Total number of gains: 97; Total number of losses: 70; Total Number of normals: 321.
Somatic mutations of MLC1:
Generating mutation plots.
Highly correlated genes for MLC1:
Showing top 20/166 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MLC1 | NPBWR1 | 0.712872 | 3 | 0 | 3 |
| MLC1 | AAAS | 0.695131 | 3 | 0 | 3 |
| MLC1 | MAPK8IP2 | 0.659664 | 4 | 0 | 4 |
| MLC1 | PLG | 0.646768 | 3 | 0 | 3 |
| MLC1 | IQSEC2 | 0.643938 | 3 | 0 | 3 |
| MLC1 | NEUROD4 | 0.640817 | 4 | 0 | 3 |
| MLC1 | HMX1 | 0.639706 | 3 | 0 | 3 |
| MLC1 | MAGEB4 | 0.634057 | 3 | 0 | 3 |
| MLC1 | CLSTN3 | 0.63357 | 3 | 0 | 3 |
| MLC1 | TMEM59L | 0.632831 | 5 | 0 | 5 |
| MLC1 | GRID2 | 0.632347 | 4 | 0 | 4 |
| MLC1 | ARVCF | 0.630367 | 5 | 0 | 5 |
| MLC1 | SNAPC2 | 0.627278 | 4 | 0 | 3 |
| MLC1 | GJA4 | 0.626444 | 4 | 0 | 3 |
| MLC1 | FMNL1 | 0.625934 | 3 | 0 | 3 |
| MLC1 | CUL7 | 0.622062 | 3 | 0 | 3 |
| MLC1 | PRR34 | 0.620741 | 4 | 0 | 3 |
| MLC1 | GDF2 | 0.617582 | 4 | 0 | 4 |
| MLC1 | HTR1A | 0.616801 | 3 | 0 | 3 |
| MLC1 | PTPRU | 0.615124 | 3 | 0 | 3 |
For details and further investigation, click here