| Full name: matrix metallopeptidase 27 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 11q22.2 | ||
| Entrez ID: 64066 | HGNC ID: HGNC:14250 | Ensembl Gene: ENSG00000137675 | OMIM ID: 618101 |
| Drug and gene relationship at DGIdb | |||
Expression of MMP27:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MMP27 | 64066 | 220783_at | -0.1760 | 0.4961 | |
| GSE20347 | MMP27 | 64066 | 220783_at | 0.1628 | 0.0520 | |
| GSE23400 | MMP27 | 64066 | 220783_at | -0.0588 | 0.0182 | |
| GSE26886 | MMP27 | 64066 | 220783_at | -0.0150 | 0.8643 | |
| GSE29001 | MMP27 | 64066 | 220783_at | -0.0827 | 0.6851 | |
| GSE38129 | MMP27 | 64066 | 220783_at | -0.0058 | 0.9516 | |
| GSE45670 | MMP27 | 64066 | 220783_at | -0.1543 | 0.3868 | |
| GSE53622 | MMP27 | 64066 | 62466 | -0.6275 | 0.0034 | |
| GSE53624 | MMP27 | 64066 | 62466 | -1.1871 | 0.0000 | |
| GSE63941 | MMP27 | 64066 | 220783_at | -0.3984 | 0.0097 | |
| GSE77861 | MMP27 | 64066 | 220783_at | -0.0387 | 0.7572 | |
| TCGA | MMP27 | 64066 | RNAseq | -2.5885 | 0.0158 |
Upregulated datasets: 0; Downregulated datasets: 2.
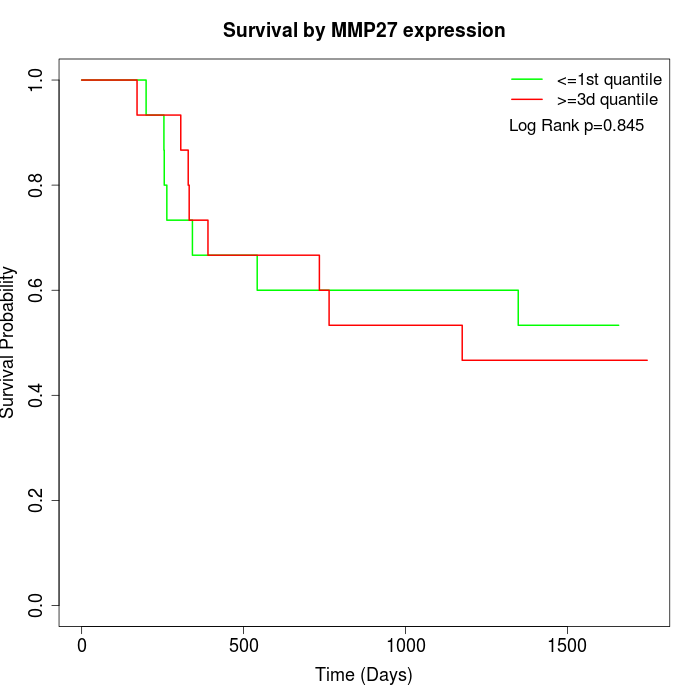
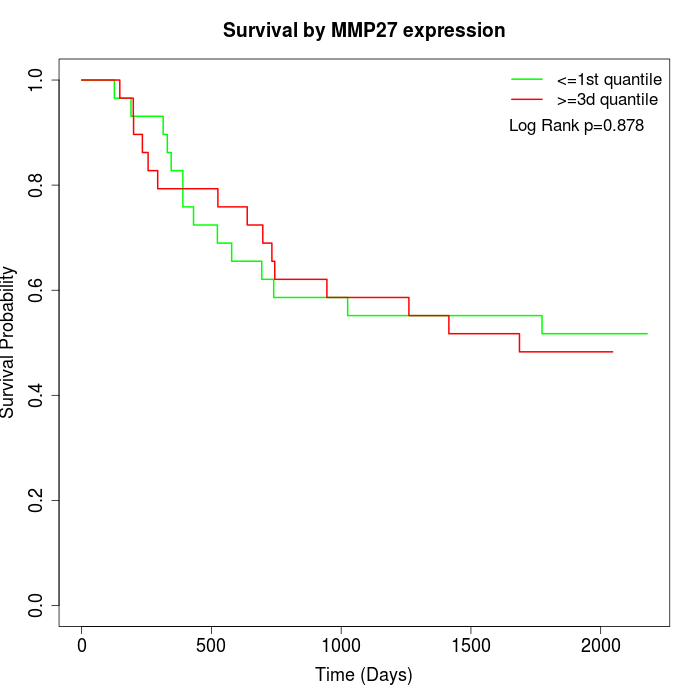
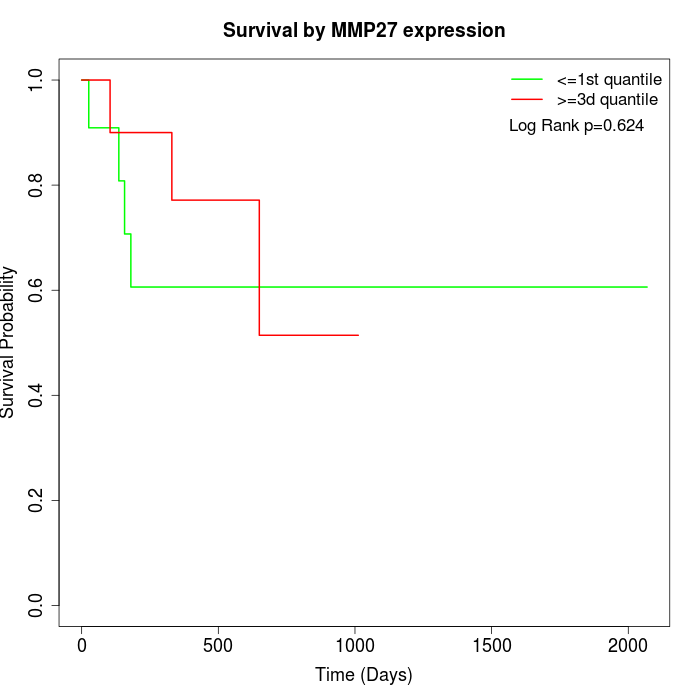
Survival by MMP27 expression:
Note: Click image to view full size file.
Copy number change of MMP27:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MMP27 | 64066 | 5 | 8 | 17 | |
| GSE20123 | MMP27 | 64066 | 5 | 8 | 17 | |
| GSE43470 | MMP27 | 64066 | 5 | 5 | 33 | |
| GSE46452 | MMP27 | 64066 | 5 | 24 | 30 | |
| GSE47630 | MMP27 | 64066 | 2 | 19 | 19 | |
| GSE54993 | MMP27 | 64066 | 9 | 2 | 59 | |
| GSE54994 | MMP27 | 64066 | 6 | 18 | 29 | |
| GSE60625 | MMP27 | 64066 | 0 | 3 | 8 | |
| GSE74703 | MMP27 | 64066 | 3 | 4 | 29 | |
| GSE74704 | MMP27 | 64066 | 5 | 4 | 11 | |
| TCGA | MMP27 | 64066 | 10 | 44 | 42 |
Total number of gains: 55; Total number of losses: 139; Total Number of normals: 294.
Somatic mutations of MMP27:
Generating mutation plots.
Highly correlated genes for MMP27:
Showing top 20/89 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MMP27 | MAP4 | 0.67999 | 3 | 0 | 3 |
| MMP27 | MAGEL2 | 0.65289 | 4 | 0 | 4 |
| MMP27 | OPRM1 | 0.639777 | 3 | 0 | 3 |
| MMP27 | LRCH1 | 0.631839 | 4 | 0 | 3 |
| MMP27 | CNTN5 | 0.630616 | 4 | 0 | 3 |
| MMP27 | TSHB | 0.616675 | 4 | 0 | 3 |
| MMP27 | PITPNM3 | 0.613257 | 4 | 0 | 3 |
| MMP27 | COL4A3 | 0.606951 | 4 | 0 | 3 |
| MMP27 | OR10H3 | 0.604213 | 4 | 0 | 3 |
| MMP27 | MTMR9 | 0.601442 | 3 | 0 | 3 |
| MMP27 | GABRA2 | 0.60119 | 4 | 0 | 4 |
| MMP27 | ATG10 | 0.599108 | 5 | 0 | 4 |
| MMP27 | ATP6V1G2 | 0.594725 | 6 | 0 | 4 |
| MMP27 | SST | 0.593234 | 4 | 0 | 3 |
| MMP27 | CLCF1 | 0.591481 | 3 | 0 | 3 |
| MMP27 | AK5 | 0.585395 | 4 | 0 | 3 |
| MMP27 | S100A5 | 0.579584 | 4 | 0 | 3 |
| MMP27 | ABCA6 | 0.578498 | 5 | 0 | 3 |
| MMP27 | CMA1 | 0.573634 | 4 | 0 | 3 |
| MMP27 | HIPK3 | 0.572987 | 6 | 0 | 4 |
For details and further investigation, click here