| Full name: mutS homolog 4 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1p31.1 | ||
| Entrez ID: 4438 | HGNC ID: HGNC:7327 | Ensembl Gene: ENSG00000057468 | OMIM ID: 602105 |
| Drug and gene relationship at DGIdb | |||
Expression of MSH4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MSH4 | 4438 | 210533_at | -0.2493 | 0.3841 | |
| GSE20347 | MSH4 | 4438 | 210533_at | -0.0831 | 0.1172 | |
| GSE23400 | MSH4 | 4438 | 210533_at | -0.0970 | 0.0000 | |
| GSE26886 | MSH4 | 4438 | 210533_at | -0.0606 | 0.4524 | |
| GSE29001 | MSH4 | 4438 | 210533_at | -0.0657 | 0.6726 | |
| GSE38129 | MSH4 | 4438 | 210533_at | -0.1168 | 0.0219 | |
| GSE45670 | MSH4 | 4438 | 210533_at | -0.0790 | 0.5102 | |
| GSE53622 | MSH4 | 4438 | 63070 | 0.0016 | 0.9910 | |
| GSE53624 | MSH4 | 4438 | 63070 | -0.0525 | 0.6143 | |
| GSE63941 | MSH4 | 4438 | 210533_at | -0.1050 | 0.4794 | |
| GSE77861 | MSH4 | 4438 | 210533_at | -0.1331 | 0.0946 | |
| TCGA | MSH4 | 4438 | RNAseq | -0.6086 | 0.4703 |
Upregulated datasets: 0; Downregulated datasets: 0.
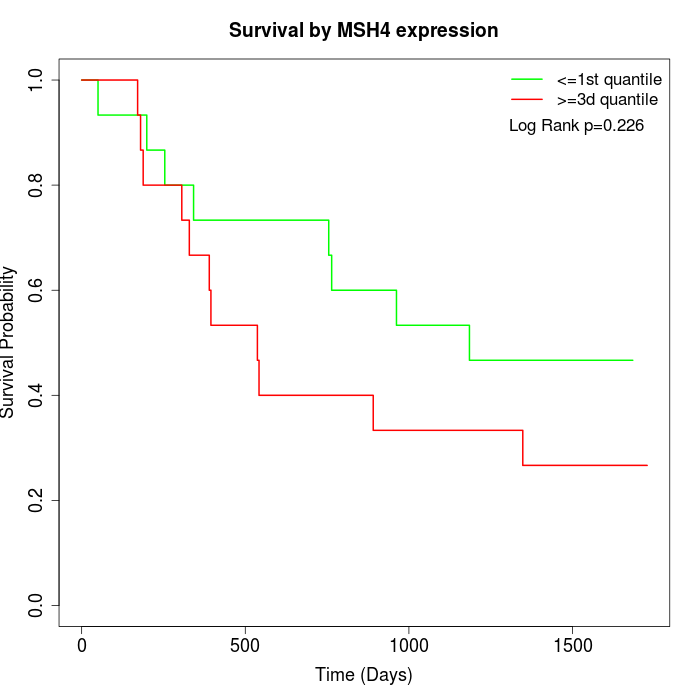
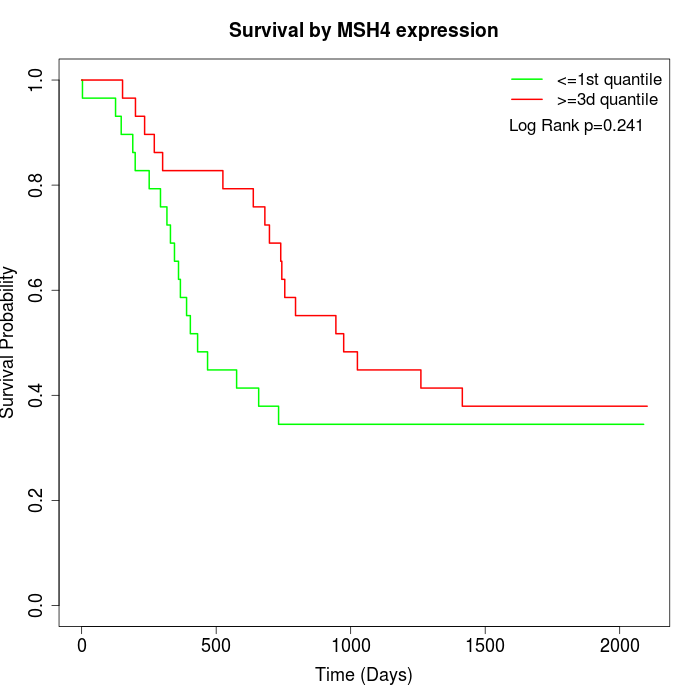
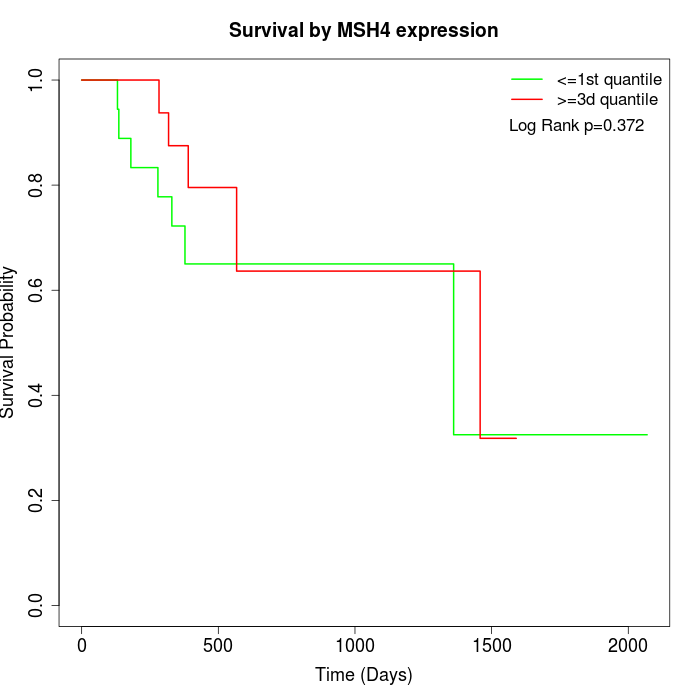
Survival by MSH4 expression:
Note: Click image to view full size file.
Copy number change of MSH4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MSH4 | 4438 | 0 | 7 | 23 | |
| GSE20123 | MSH4 | 4438 | 1 | 7 | 22 | |
| GSE43470 | MSH4 | 4438 | 3 | 4 | 36 | |
| GSE46452 | MSH4 | 4438 | 0 | 1 | 58 | |
| GSE47630 | MSH4 | 4438 | 8 | 6 | 26 | |
| GSE54993 | MSH4 | 4438 | 0 | 0 | 70 | |
| GSE54994 | MSH4 | 4438 | 6 | 3 | 44 | |
| GSE60625 | MSH4 | 4438 | 0 | 0 | 11 | |
| GSE74703 | MSH4 | 4438 | 2 | 3 | 31 | |
| GSE74704 | MSH4 | 4438 | 1 | 4 | 15 | |
| TCGA | MSH4 | 4438 | 8 | 24 | 64 |
Total number of gains: 29; Total number of losses: 59; Total Number of normals: 400.
Somatic mutations of MSH4:
Generating mutation plots.
Highly correlated genes for MSH4:
Showing top 20/564 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MSH4 | CIITA | 0.746948 | 3 | 0 | 3 |
| MSH4 | PRDM11 | 0.729364 | 3 | 0 | 3 |
| MSH4 | ADRB3 | 0.712728 | 3 | 0 | 3 |
| MSH4 | GH2 | 0.699395 | 4 | 0 | 4 |
| MSH4 | APOBEC1 | 0.698107 | 4 | 0 | 4 |
| MSH4 | DCLK2 | 0.694222 | 4 | 0 | 4 |
| MSH4 | ASIC4 | 0.690601 | 3 | 0 | 3 |
| MSH4 | APBA1 | 0.690542 | 3 | 0 | 3 |
| MSH4 | SSTR4 | 0.685191 | 5 | 0 | 4 |
| MSH4 | IL25 | 0.682123 | 3 | 0 | 3 |
| MSH4 | CAMK4 | 0.681956 | 3 | 0 | 3 |
| MSH4 | CDKL5 | 0.679876 | 4 | 0 | 4 |
| MSH4 | AGTR2 | 0.677613 | 4 | 0 | 3 |
| MSH4 | SYT5 | 0.676042 | 3 | 0 | 3 |
| MSH4 | SLC17A3 | 0.674404 | 3 | 0 | 3 |
| MSH4 | PSG2 | 0.670551 | 4 | 0 | 3 |
| MSH4 | ULK4 | 0.670313 | 3 | 0 | 3 |
| MSH4 | PPM1E | 0.667786 | 5 | 0 | 5 |
| MSH4 | PGC | 0.666985 | 3 | 0 | 3 |
| MSH4 | KIR2DS3 | 0.666254 | 3 | 0 | 3 |
For details and further investigation, click here