| Full name: mitochondrial transcription termination factor 4 | Alias Symbol: MGC61716 | ||
| Type: protein-coding gene | Cytoband: 2q37.3 | ||
| Entrez ID: 130916 | HGNC ID: HGNC:28785 | Ensembl Gene: ENSG00000122085 | OMIM ID: 615393 |
| Drug and gene relationship at DGIdb | |||
Expression of MTERF4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MTERF4 | 130916 | 1557966_x_at | -0.6568 | 0.1353 | |
| GSE20347 | MTERF4 | 130916 | 214364_at | -1.2561 | 0.0000 | |
| GSE23400 | MTERF4 | 130916 | 214364_at | -0.4858 | 0.0000 | |
| GSE26886 | MTERF4 | 130916 | 1557966_x_at | -0.7904 | 0.0010 | |
| GSE29001 | MTERF4 | 130916 | 214364_at | -1.1816 | 0.0000 | |
| GSE38129 | MTERF4 | 130916 | 214364_at | -1.0130 | 0.0000 | |
| GSE45670 | MTERF4 | 130916 | 226486_at | -0.5674 | 0.0134 | |
| GSE53622 | MTERF4 | 130916 | 18144 | -0.2686 | 0.0055 | |
| GSE53624 | MTERF4 | 130916 | 55610 | -0.4998 | 0.0000 | |
| GSE63941 | MTERF4 | 130916 | 1557965_at | -0.7198 | 0.1283 | |
| GSE77861 | MTERF4 | 130916 | 1557965_at | -0.4875 | 0.1114 | |
| SRP007169 | MTERF4 | 130916 | RNAseq | -1.7278 | 0.0000 | |
| SRP008496 | MTERF4 | 130916 | RNAseq | -1.7333 | 0.0000 | |
| SRP064894 | MTERF4 | 130916 | RNAseq | -0.9880 | 0.0000 | |
| SRP133303 | MTERF4 | 130916 | RNAseq | -0.8218 | 0.0000 | |
| SRP159526 | MTERF4 | 130916 | RNAseq | -0.8683 | 0.0008 | |
| SRP193095 | MTERF4 | 130916 | RNAseq | -1.0411 | 0.0000 | |
| SRP219564 | MTERF4 | 130916 | RNAseq | -0.9168 | 0.0070 |
Upregulated datasets: 0; Downregulated datasets: 6.
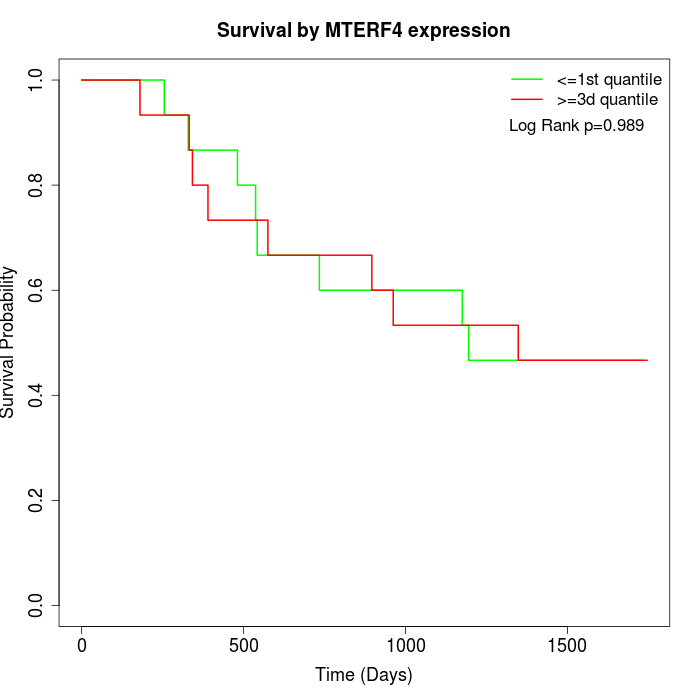
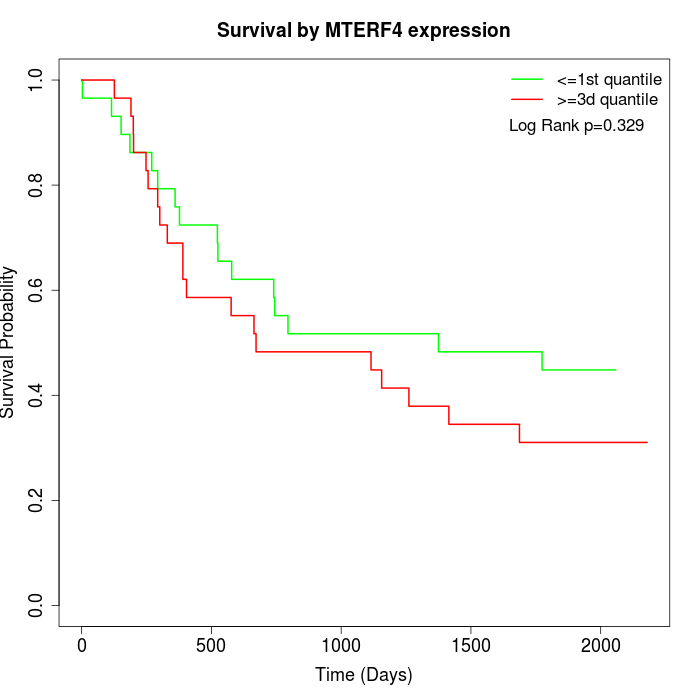
Survival by MTERF4 expression:
Note: Click image to view full size file.
Copy number change of MTERF4:
No record found for this gene.
Somatic mutations of MTERF4:
Generating mutation plots.
Highly correlated genes for MTERF4:
Showing top 20/1095 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MTERF4 | CNPPD1 | 0.780451 | 8 | 0 | 8 |
| MTERF4 | MIR210HG | 0.752473 | 3 | 0 | 3 |
| MTERF4 | PPP1R7 | 0.723749 | 11 | 0 | 11 |
| MTERF4 | MXD1 | 0.719959 | 9 | 0 | 7 |
| MTERF4 | EPB41L4A | 0.718556 | 8 | 0 | 7 |
| MTERF4 | HECTD3 | 0.712647 | 3 | 0 | 3 |
| MTERF4 | IL17RC | 0.711942 | 6 | 0 | 5 |
| MTERF4 | RIOK3 | 0.710813 | 8 | 0 | 8 |
| MTERF4 | ACOX1 | 0.709237 | 8 | 0 | 8 |
| MTERF4 | CYP2C18 | 0.706377 | 7 | 0 | 7 |
| MTERF4 | HDHD2 | 0.705528 | 3 | 0 | 3 |
| MTERF4 | SORT1 | 0.704794 | 9 | 0 | 8 |
| MTERF4 | PADI1 | 0.704183 | 8 | 0 | 7 |
| MTERF4 | GYS2 | 0.703318 | 10 | 0 | 8 |
| MTERF4 | SIRT7 | 0.702046 | 8 | 0 | 8 |
| MTERF4 | TJP3 | 0.701592 | 8 | 0 | 8 |
| MTERF4 | EPB41L3 | 0.701477 | 9 | 0 | 8 |
| MTERF4 | SPAG16 | 0.70127 | 9 | 0 | 7 |
| MTERF4 | CEACAM5 | 0.699925 | 7 | 0 | 7 |
| MTERF4 | ABLIM1 | 0.699521 | 8 | 0 | 7 |
For details and further investigation, click here