| Full name: melatonin receptor 1A | Alias Symbol: MEL-1A-R | ||
| Type: protein-coding gene | Cytoband: 4q35.2 | ||
| Entrez ID: 4543 | HGNC ID: HGNC:7463 | Ensembl Gene: ENSG00000168412 | OMIM ID: 600665 |
| Drug and gene relationship at DGIdb | |||
MTNR1A involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04713 | Circadian entrainment |
Expression of MTNR1A:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MTNR1A | 4543 | 221369_at | 0.0497 | 0.8423 | |
| GSE20347 | MTNR1A | 4543 | 221369_at | -0.0824 | 0.1771 | |
| GSE23400 | MTNR1A | 4543 | 221369_at | -0.0652 | 0.0124 | |
| GSE26886 | MTNR1A | 4543 | 221369_at | -0.1409 | 0.1438 | |
| GSE29001 | MTNR1A | 4543 | 221369_at | 0.0321 | 0.7996 | |
| GSE38129 | MTNR1A | 4543 | 221369_at | -0.0855 | 0.2510 | |
| GSE45670 | MTNR1A | 4543 | 221369_at | 0.0908 | 0.2782 | |
| GSE63941 | MTNR1A | 4543 | 221369_at | 0.0957 | 0.4802 | |
| GSE77861 | MTNR1A | 4543 | 221369_at | -0.0369 | 0.6834 | |
| GSE97050 | MTNR1A | 4543 | A_24_P266019 | 0.3991 | 0.1801 | |
| TCGA | MTNR1A | 4543 | RNAseq | 0.3525 | 0.6697 |
Upregulated datasets: 0; Downregulated datasets: 0.
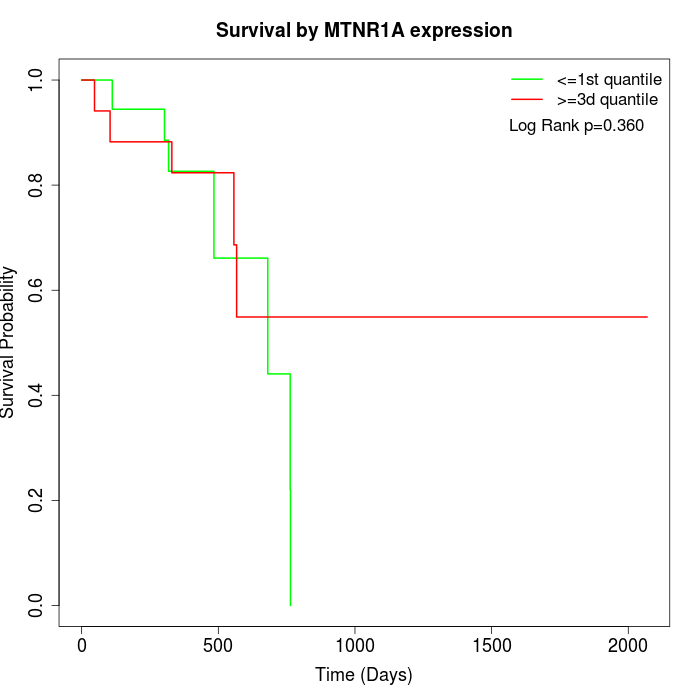
Survival by MTNR1A expression:
Note: Click image to view full size file.
Copy number change of MTNR1A:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MTNR1A | 4543 | 2 | 12 | 16 | |
| GSE20123 | MTNR1A | 4543 | 2 | 12 | 16 | |
| GSE43470 | MTNR1A | 4543 | 1 | 15 | 27 | |
| GSE46452 | MTNR1A | 4543 | 1 | 36 | 22 | |
| GSE47630 | MTNR1A | 4543 | 0 | 22 | 18 | |
| GSE54993 | MTNR1A | 4543 | 10 | 0 | 60 | |
| GSE54994 | MTNR1A | 4543 | 2 | 14 | 37 | |
| GSE60625 | MTNR1A | 4543 | 0 | 0 | 11 | |
| GSE74703 | MTNR1A | 4543 | 1 | 12 | 23 | |
| GSE74704 | MTNR1A | 4543 | 0 | 6 | 14 | |
| TCGA | MTNR1A | 4543 | 10 | 34 | 52 |
Total number of gains: 29; Total number of losses: 163; Total Number of normals: 296.
Somatic mutations of MTNR1A:
Generating mutation plots.
Highly correlated genes for MTNR1A:
Showing top 20/298 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MTNR1A | GTF2H3 | 0.732417 | 3 | 0 | 3 |
| MTNR1A | GUCY1A2 | 0.717213 | 3 | 0 | 3 |
| MTNR1A | KDM6B | 0.698493 | 3 | 0 | 3 |
| MTNR1A | ADAM11 | 0.666885 | 4 | 0 | 4 |
| MTNR1A | CTRB2 | 0.663536 | 3 | 0 | 3 |
| MTNR1A | IL12RB1 | 0.655055 | 4 | 0 | 4 |
| MTNR1A | QPCTL | 0.638648 | 5 | 0 | 4 |
| MTNR1A | MESP1 | 0.63501 | 3 | 0 | 3 |
| MTNR1A | INTS6 | 0.634355 | 4 | 0 | 3 |
| MTNR1A | UNC13D | 0.630537 | 3 | 0 | 3 |
| MTNR1A | SARDH | 0.627554 | 4 | 0 | 4 |
| MTNR1A | VPS53 | 0.623346 | 4 | 0 | 4 |
| MTNR1A | NPAP1 | 0.621801 | 3 | 0 | 3 |
| MTNR1A | ERCC4 | 0.619726 | 4 | 0 | 3 |
| MTNR1A | IRX3 | 0.617762 | 3 | 0 | 3 |
| MTNR1A | PTTG2 | 0.617092 | 3 | 0 | 3 |
| MTNR1A | IKZF3 | 0.616116 | 4 | 0 | 3 |
| MTNR1A | MRPS22 | 0.614151 | 4 | 0 | 3 |
| MTNR1A | HAUS2 | 0.611649 | 4 | 0 | 3 |
| MTNR1A | IRS4 | 0.611247 | 4 | 0 | 3 |
For details and further investigation, click here