| Full name: myosin heavy chain 10 | Alias Symbol: NMMHCB | ||
| Type: protein-coding gene | Cytoband: 17p13.1 | ||
| Entrez ID: 4628 | HGNC ID: HGNC:7568 | Ensembl Gene: ENSG00000133026 | OMIM ID: 160776 |
| Drug and gene relationship at DGIdb | |||
MYH10 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04530 | Tight junction | |
| hsa04810 | Regulation of actin cytoskeleton | |
| hsa05132 | Salmonella infection |
Expression of MYH10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MYH10 | 4628 | 212372_at | 1.2068 | 0.0616 | |
| GSE20347 | MYH10 | 4628 | 212372_at | 1.2178 | 0.0006 | |
| GSE23400 | MYH10 | 4628 | 212372_at | 1.0566 | 0.0000 | |
| GSE26886 | MYH10 | 4628 | 212372_at | 2.1795 | 0.0000 | |
| GSE29001 | MYH10 | 4628 | 212372_at | 1.3149 | 0.0003 | |
| GSE38129 | MYH10 | 4628 | 212372_at | 0.9886 | 0.0001 | |
| GSE45670 | MYH10 | 4628 | 212372_at | 0.4900 | 0.1992 | |
| GSE53622 | MYH10 | 4628 | 86478 | 1.0811 | 0.0000 | |
| GSE53624 | MYH10 | 4628 | 53831 | 1.0851 | 0.0000 | |
| GSE63941 | MYH10 | 4628 | 212372_at | -1.1192 | 0.0633 | |
| GSE77861 | MYH10 | 4628 | 212372_at | 1.1118 | 0.2200 | |
| GSE97050 | MYH10 | 4628 | A_33_P3235004 | 0.1413 | 0.6352 | |
| SRP007169 | MYH10 | 4628 | RNAseq | 3.0653 | 0.0000 | |
| SRP008496 | MYH10 | 4628 | RNAseq | 2.8851 | 0.0000 | |
| SRP064894 | MYH10 | 4628 | RNAseq | 2.0248 | 0.0000 | |
| SRP133303 | MYH10 | 4628 | RNAseq | 1.0203 | 0.0004 | |
| SRP159526 | MYH10 | 4628 | RNAseq | 1.4977 | 0.0000 | |
| SRP193095 | MYH10 | 4628 | RNAseq | 1.5601 | 0.0000 | |
| SRP219564 | MYH10 | 4628 | RNAseq | 1.1207 | 0.0180 | |
| TCGA | MYH10 | 4628 | RNAseq | 0.1915 | 0.0233 |
Upregulated datasets: 13; Downregulated datasets: 0.
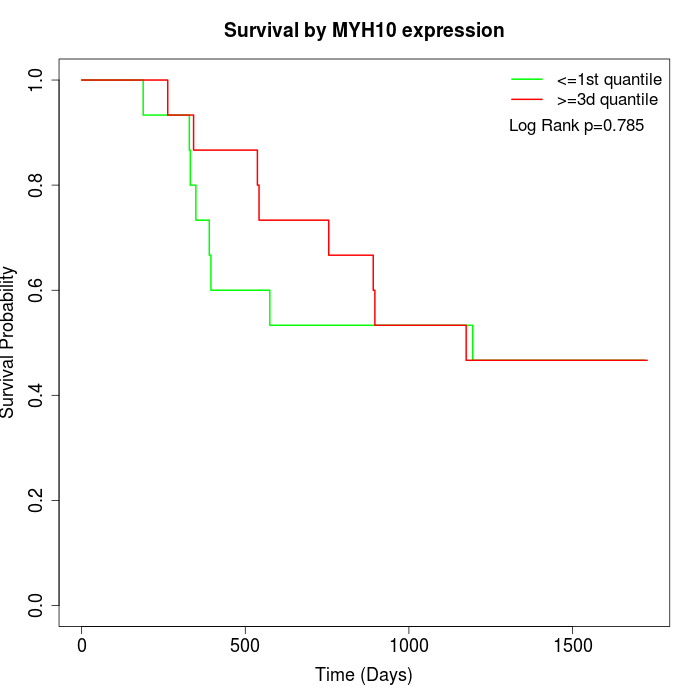
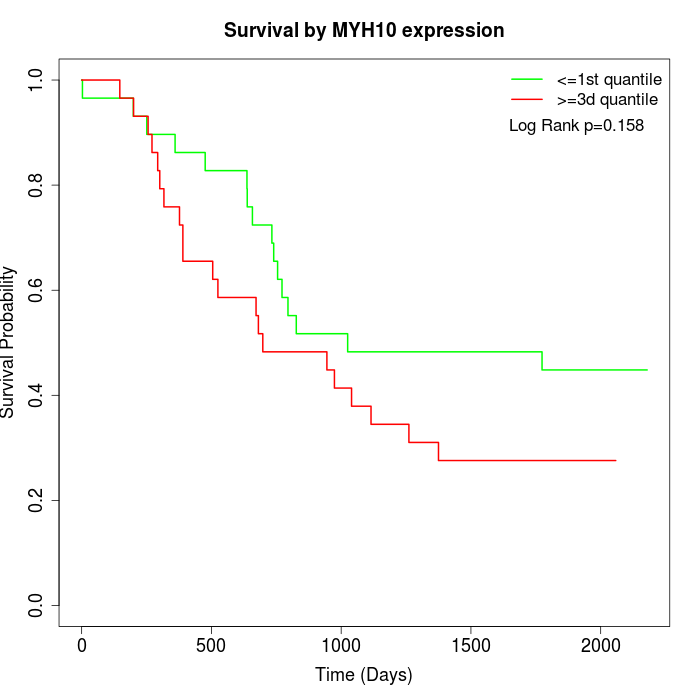
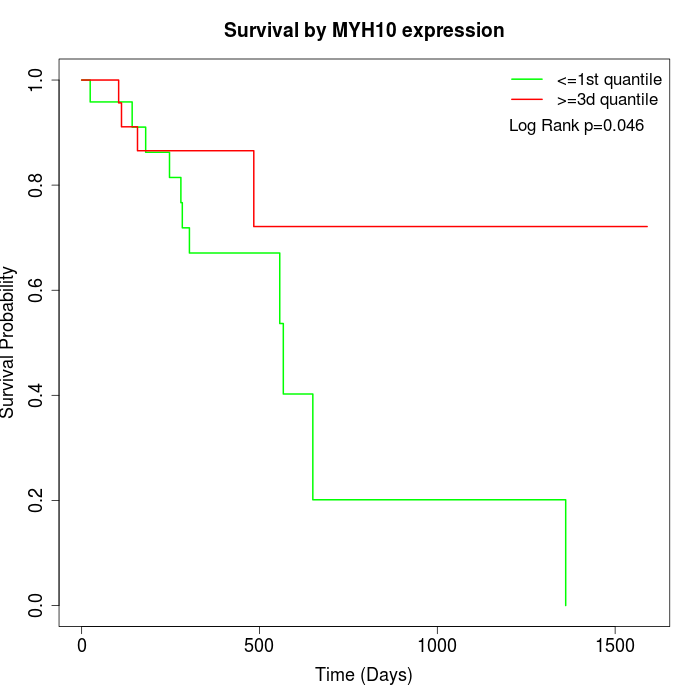
Survival by MYH10 expression:
Note: Click image to view full size file.
Copy number change of MYH10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MYH10 | 4628 | 4 | 2 | 24 | |
| GSE20123 | MYH10 | 4628 | 4 | 3 | 23 | |
| GSE43470 | MYH10 | 4628 | 1 | 6 | 36 | |
| GSE46452 | MYH10 | 4628 | 34 | 1 | 24 | |
| GSE47630 | MYH10 | 4628 | 7 | 1 | 32 | |
| GSE54993 | MYH10 | 4628 | 4 | 3 | 63 | |
| GSE54994 | MYH10 | 4628 | 5 | 8 | 40 | |
| GSE60625 | MYH10 | 4628 | 4 | 0 | 7 | |
| GSE74703 | MYH10 | 4628 | 1 | 3 | 32 | |
| GSE74704 | MYH10 | 4628 | 2 | 1 | 17 | |
| TCGA | MYH10 | 4628 | 18 | 20 | 58 |
Total number of gains: 84; Total number of losses: 48; Total Number of normals: 356.
Somatic mutations of MYH10:
Generating mutation plots.
Highly correlated genes for MYH10:
Showing top 20/1503 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MYH10 | RAB34 | 0.788658 | 3 | 0 | 3 |
| MYH10 | SLC46A1 | 0.771509 | 3 | 0 | 3 |
| MYH10 | PHRF1 | 0.759219 | 3 | 0 | 3 |
| MYH10 | CAMKK1 | 0.73818 | 4 | 0 | 4 |
| MYH10 | POLR2A | 0.73664 | 3 | 0 | 3 |
| MYH10 | SLC19A2 | 0.735091 | 3 | 0 | 3 |
| MYH10 | SLC39A13 | 0.731373 | 5 | 0 | 5 |
| MYH10 | PDE7A | 0.730981 | 6 | 0 | 6 |
| MYH10 | ROMO1 | 0.730973 | 3 | 0 | 3 |
| MYH10 | SPON2 | 0.723181 | 3 | 0 | 3 |
| MYH10 | NF1 | 0.722964 | 3 | 0 | 3 |
| MYH10 | TP53I13 | 0.71945 | 6 | 0 | 6 |
| MYH10 | SIX5 | 0.716893 | 4 | 0 | 4 |
| MYH10 | FMNL2 | 0.713432 | 7 | 0 | 6 |
| MYH10 | SPAG7 | 0.711882 | 3 | 0 | 3 |
| MYH10 | PLEKHG4B | 0.71001 | 4 | 0 | 4 |
| MYH10 | MNT | 0.707156 | 4 | 0 | 4 |
| MYH10 | LPCAT1 | 0.706333 | 11 | 0 | 10 |
| MYH10 | ALKBH6 | 0.703526 | 4 | 0 | 4 |
| MYH10 | ENAH | 0.703159 | 12 | 0 | 10 |
For details and further investigation, click here