| Full name: myosin light chain 6 | Alias Symbol: ESMLC|MLC3NM|MLC1SM | ||
| Type: protein-coding gene | Cytoband: 12q13.2 | ||
| Entrez ID: 4637 | HGNC ID: HGNC:7587 | Ensembl Gene: ENSG00000092841 | OMIM ID: 609931 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
MYL6 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04270 | Vascular smooth muscle contraction | |
| hsa04921 | Oxytocin signaling pathway |
Expression of MYL6:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MYL6 | 4637 | 212082_s_at | -0.2747 | 0.2840 | |
| GSE20347 | MYL6 | 4637 | 212082_s_at | -0.4776 | 0.0004 | |
| GSE23400 | MYL6 | 4637 | 212082_s_at | -0.3376 | 0.0000 | |
| GSE26886 | MYL6 | 4637 | 212082_s_at | -0.6023 | 0.0006 | |
| GSE29001 | MYL6 | 4637 | 212082_s_at | -0.5650 | 0.1197 | |
| GSE38129 | MYL6 | 4637 | 212082_s_at | -0.4572 | 0.0028 | |
| GSE45670 | MYL6 | 4637 | 212082_s_at | -0.1826 | 0.0274 | |
| GSE53622 | MYL6 | 4637 | 67773 | -0.9356 | 0.0000 | |
| GSE53624 | MYL6 | 4637 | 67773 | -0.4822 | 0.0000 | |
| GSE63941 | MYL6 | 4637 | 212082_s_at | -0.8267 | 0.0976 | |
| GSE77861 | MYL6 | 4637 | 212082_s_at | -0.3215 | 0.0166 | |
| GSE97050 | MYL6 | 4637 | A_24_P56130 | -1.0510 | 0.1009 | |
| SRP007169 | MYL6 | 4637 | RNAseq | -1.1731 | 0.0004 | |
| SRP008496 | MYL6 | 4637 | RNAseq | -0.8358 | 0.0001 | |
| SRP064894 | MYL6 | 4637 | RNAseq | -0.6280 | 0.0257 | |
| SRP133303 | MYL6 | 4637 | RNAseq | -0.1968 | 0.3191 | |
| SRP159526 | MYL6 | 4637 | RNAseq | -0.8228 | 0.0003 | |
| SRP193095 | MYL6 | 4637 | RNAseq | -0.6857 | 0.0000 | |
| SRP219564 | MYL6 | 4637 | RNAseq | -0.9442 | 0.1303 | |
| TCGA | MYL6 | 4637 | RNAseq | -0.1085 | 0.0142 |
Upregulated datasets: 0; Downregulated datasets: 1.
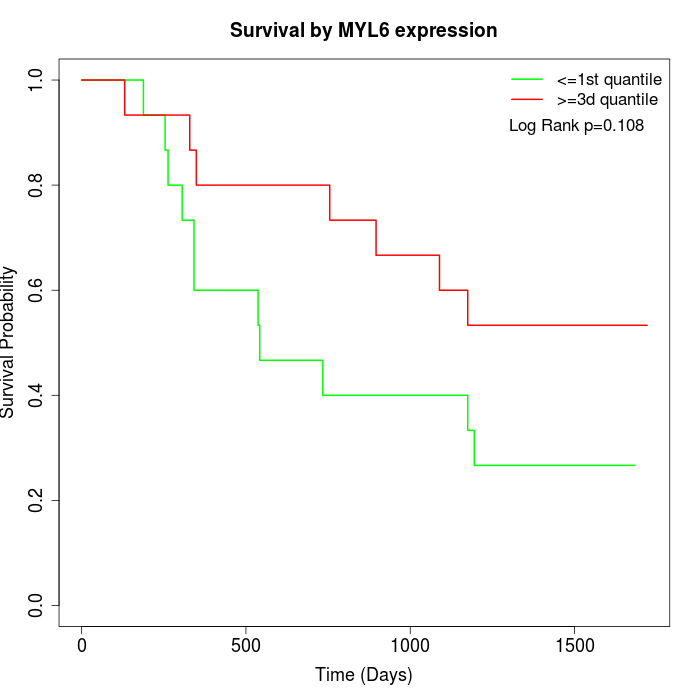
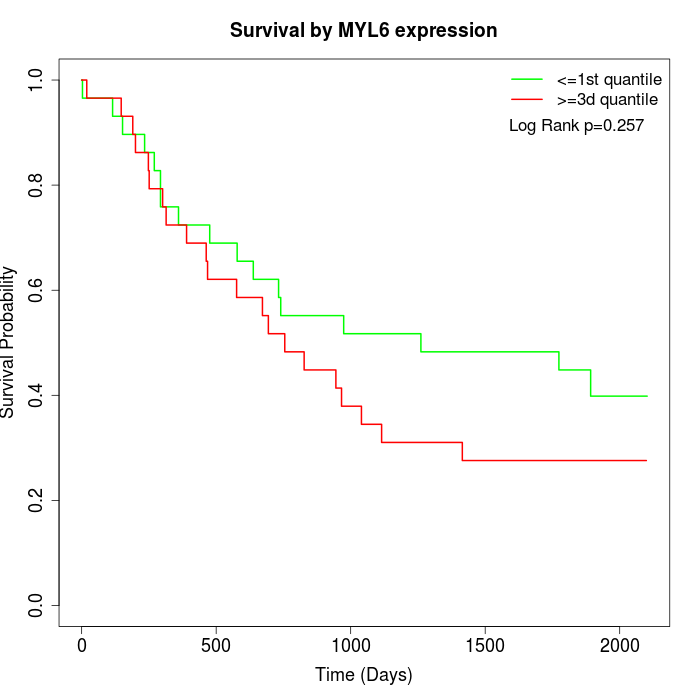
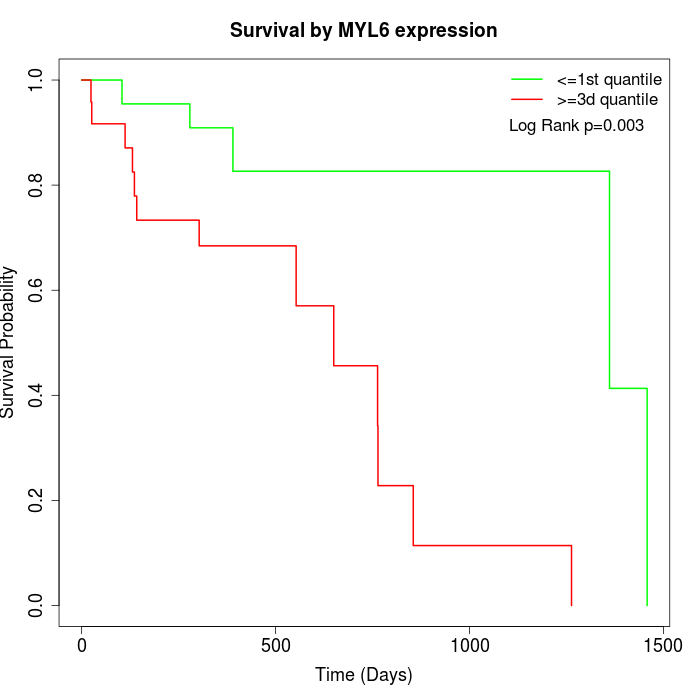
Survival by MYL6 expression:
Note: Click image to view full size file.
Copy number change of MYL6:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MYL6 | 4637 | 7 | 2 | 21 | |
| GSE20123 | MYL6 | 4637 | 7 | 1 | 22 | |
| GSE43470 | MYL6 | 4637 | 4 | 0 | 39 | |
| GSE46452 | MYL6 | 4637 | 8 | 1 | 50 | |
| GSE47630 | MYL6 | 4637 | 10 | 2 | 28 | |
| GSE54993 | MYL6 | 4637 | 0 | 5 | 65 | |
| GSE54994 | MYL6 | 4637 | 4 | 1 | 48 | |
| GSE60625 | MYL6 | 4637 | 0 | 0 | 11 | |
| GSE74703 | MYL6 | 4637 | 4 | 0 | 32 | |
| GSE74704 | MYL6 | 4637 | 4 | 1 | 15 | |
| TCGA | MYL6 | 4637 | 14 | 10 | 72 |
Total number of gains: 62; Total number of losses: 23; Total Number of normals: 403.
Somatic mutations of MYL6:
Generating mutation plots.
Highly correlated genes for MYL6:
Showing top 20/1300 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MYL6 | ANKS1B | 0.844762 | 3 | 0 | 3 |
| MYL6 | FBXO30 | 0.824874 | 3 | 0 | 3 |
| MYL6 | SORCS1 | 0.773757 | 4 | 0 | 4 |
| MYL6 | SLMAP | 0.764125 | 7 | 0 | 7 |
| MYL6 | RAP1A | 0.756416 | 10 | 0 | 10 |
| MYL6 | PPARGC1A | 0.749846 | 5 | 0 | 5 |
| MYL6 | THRB | 0.746589 | 5 | 0 | 4 |
| MYL6 | EIF4E3 | 0.741259 | 7 | 0 | 7 |
| MYL6 | ACTBL2 | 0.739586 | 3 | 0 | 3 |
| MYL6 | SLC35F1 | 0.737568 | 3 | 0 | 3 |
| MYL6 | CADM2 | 0.735914 | 4 | 0 | 3 |
| MYL6 | CYP21A2 | 0.733717 | 3 | 0 | 3 |
| MYL6 | CLEC3B | 0.732498 | 3 | 0 | 3 |
| MYL6 | PLP1 | 0.731426 | 7 | 0 | 7 |
| MYL6 | ZCCHC9 | 0.72004 | 3 | 0 | 3 |
| MYL6 | NRXN3 | 0.714778 | 5 | 0 | 5 |
| MYL6 | C3orf70 | 0.714545 | 4 | 0 | 3 |
| MYL6 | SH3BGR | 0.714036 | 7 | 0 | 6 |
| MYL6 | ASB5 | 0.709247 | 6 | 0 | 5 |
| MYL6 | LRFN5 | 0.705879 | 4 | 0 | 3 |
For details and further investigation, click here