| Full name: neutrophil cytosolic factor 4 | Alias Symbol: p40phox|SH3PXD4 | ||
| Type: protein-coding gene | Cytoband: 22q12.3 | ||
| Entrez ID: 4689 | HGNC ID: HGNC:7662 | Ensembl Gene: ENSG00000100365 | OMIM ID: 601488 |
| Drug and gene relationship at DGIdb | |||
NCF4 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04380 | Osteoclast differentiation | |
| hsa04670 | Leukocyte transendothelial migration | |
| hsa05140 | Leishmaniasis |
Expression of NCF4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NCF4 | 4689 | 205147_x_at | 0.3316 | 0.6813 | |
| GSE20347 | NCF4 | 4689 | 205147_x_at | -0.1354 | 0.1403 | |
| GSE23400 | NCF4 | 4689 | 205147_x_at | 0.0002 | 0.9978 | |
| GSE26886 | NCF4 | 4689 | 205147_x_at | -0.3721 | 0.0514 | |
| GSE29001 | NCF4 | 4689 | 205147_x_at | -0.1465 | 0.3656 | |
| GSE38129 | NCF4 | 4689 | 205147_x_at | -0.1456 | 0.2781 | |
| GSE45670 | NCF4 | 4689 | 205147_x_at | -0.0431 | 0.8875 | |
| GSE53622 | NCF4 | 4689 | 67700 | 0.1514 | 0.1201 | |
| GSE53624 | NCF4 | 4689 | 67700 | -0.0594 | 0.4637 | |
| GSE63941 | NCF4 | 4689 | 205147_x_at | 0.3801 | 0.1225 | |
| GSE77861 | NCF4 | 4689 | 205147_x_at | -0.1256 | 0.3601 | |
| GSE97050 | NCF4 | 4689 | A_33_P3365432 | 0.5963 | 0.1399 | |
| SRP007169 | NCF4 | 4689 | RNAseq | 0.2211 | 0.7681 | |
| SRP008496 | NCF4 | 4689 | RNAseq | 0.2674 | 0.5725 | |
| SRP064894 | NCF4 | 4689 | RNAseq | 0.6662 | 0.0303 | |
| SRP133303 | NCF4 | 4689 | RNAseq | 0.0737 | 0.7914 | |
| SRP159526 | NCF4 | 4689 | RNAseq | -0.1241 | 0.8211 | |
| SRP193095 | NCF4 | 4689 | RNAseq | 0.4008 | 0.1268 | |
| SRP219564 | NCF4 | 4689 | RNAseq | 1.0824 | 0.0746 | |
| TCGA | NCF4 | 4689 | RNAseq | -0.0373 | 0.8008 |
Upregulated datasets: 0; Downregulated datasets: 0.
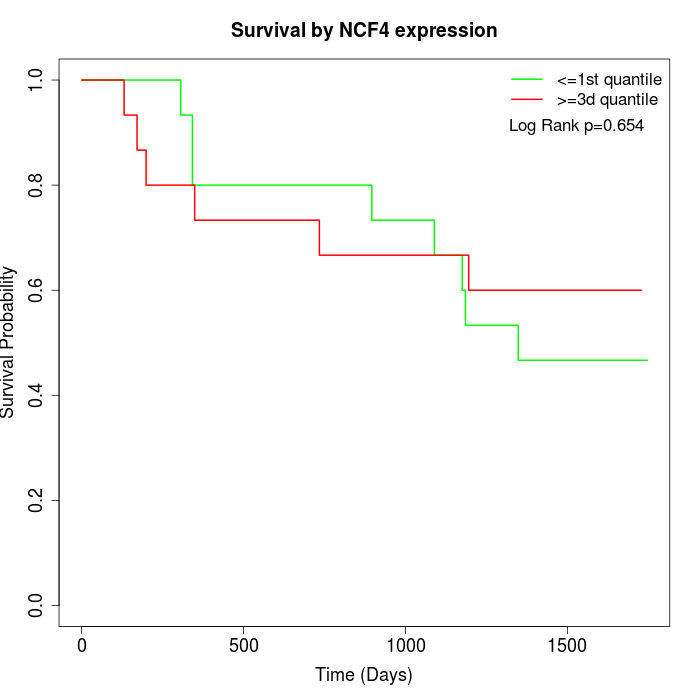
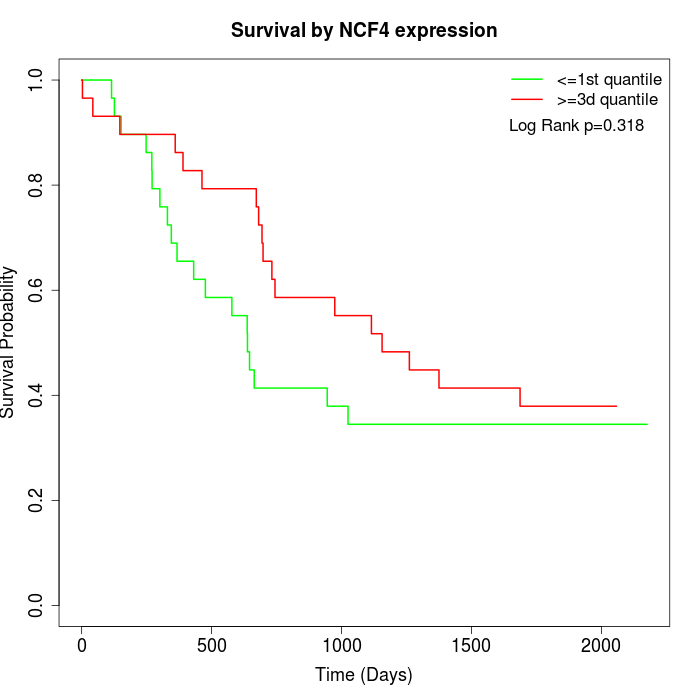
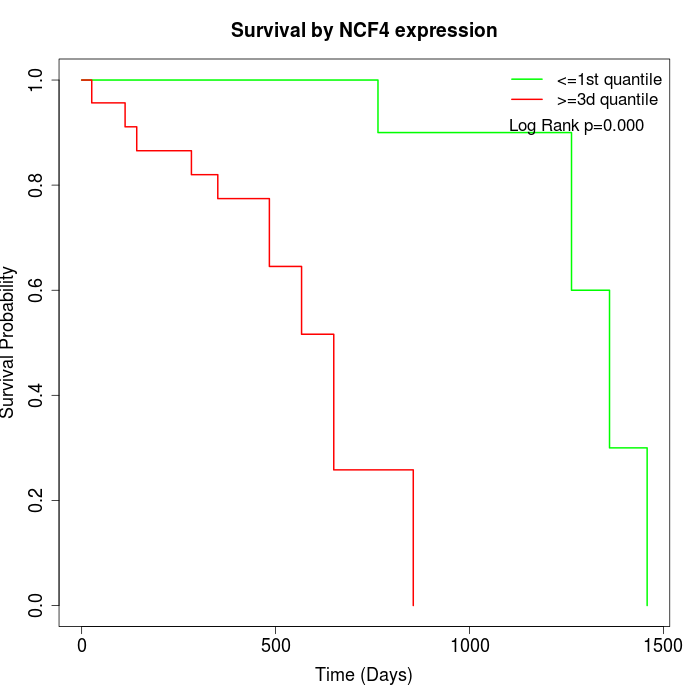
Survival by NCF4 expression:
Note: Click image to view full size file.
Copy number change of NCF4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NCF4 | 4689 | 6 | 6 | 18 | |
| GSE20123 | NCF4 | 4689 | 6 | 5 | 19 | |
| GSE43470 | NCF4 | 4689 | 4 | 6 | 33 | |
| GSE46452 | NCF4 | 4689 | 31 | 1 | 27 | |
| GSE47630 | NCF4 | 4689 | 8 | 5 | 27 | |
| GSE54993 | NCF4 | 4689 | 3 | 6 | 61 | |
| GSE54994 | NCF4 | 4689 | 10 | 9 | 34 | |
| GSE60625 | NCF4 | 4689 | 5 | 0 | 6 | |
| GSE74703 | NCF4 | 4689 | 4 | 4 | 28 | |
| GSE74704 | NCF4 | 4689 | 3 | 2 | 15 | |
| TCGA | NCF4 | 4689 | 27 | 16 | 53 |
Total number of gains: 107; Total number of losses: 60; Total Number of normals: 321.
Somatic mutations of NCF4:
Generating mutation plots.
Highly correlated genes for NCF4:
Showing top 20/263 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NCF4 | PARVG | 0.703488 | 6 | 0 | 5 |
| NCF4 | ATG13 | 0.694557 | 3 | 0 | 3 |
| NCF4 | DOCK2 | 0.684402 | 11 | 0 | 9 |
| NCF4 | CSF2RB | 0.676648 | 10 | 0 | 9 |
| NCF4 | MOCOS | 0.67209 | 3 | 0 | 3 |
| NCF4 | FERMT3 | 0.671617 | 5 | 0 | 4 |
| NCF4 | CYTIP | 0.665174 | 8 | 0 | 8 |
| NCF4 | ABI3 | 0.661612 | 6 | 0 | 6 |
| NCF4 | DOCK10 | 0.653124 | 7 | 0 | 7 |
| NCF4 | BTLA | 0.651933 | 4 | 0 | 3 |
| NCF4 | CD53 | 0.651035 | 8 | 0 | 7 |
| NCF4 | PIK3CG | 0.650344 | 7 | 0 | 6 |
| NCF4 | MS4A7 | 0.646397 | 3 | 0 | 3 |
| NCF4 | CD300C | 0.645914 | 5 | 0 | 4 |
| NCF4 | MYO1F | 0.64427 | 11 | 0 | 10 |
| NCF4 | BTK | 0.639457 | 7 | 0 | 5 |
| NCF4 | PRKCB | 0.635748 | 6 | 0 | 6 |
| NCF4 | ARHGAP9 | 0.635019 | 6 | 0 | 4 |
| NCF4 | PTPRC | 0.63251 | 10 | 0 | 7 |
| NCF4 | AKNA | 0.631096 | 6 | 0 | 5 |
For details and further investigation, click here