| Full name: NCK adaptor protein 1 | Alias Symbol: NCKalpha | ||
| Type: protein-coding gene | Cytoband: 3q22.3 | ||
| Entrez ID: 4690 | HGNC ID: HGNC:7664 | Ensembl Gene: ENSG00000158092 | OMIM ID: 600508 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
NCK1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04012 | ErbB signaling pathway | |
| hsa04360 | Axon guidance | |
| hsa04660 | T cell receptor signaling pathway | |
| hsa05130 | Pathogenic Escherichia coli infection |
Expression of NCK1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NCK1 | 4690 | 211063_s_at | 0.0304 | 0.9518 | |
| GSE20347 | NCK1 | 4690 | 211063_s_at | 0.0193 | 0.9286 | |
| GSE23400 | NCK1 | 4690 | 204725_s_at | -0.1660 | 0.2257 | |
| GSE26886 | NCK1 | 4690 | 211063_s_at | -0.5834 | 0.0204 | |
| GSE29001 | NCK1 | 4690 | 211063_s_at | 0.0323 | 0.9176 | |
| GSE38129 | NCK1 | 4690 | 211063_s_at | 0.1416 | 0.5047 | |
| GSE45670 | NCK1 | 4690 | 211063_s_at | 0.0564 | 0.8076 | |
| GSE53622 | NCK1 | 4690 | 83062 | -0.0077 | 0.9473 | |
| GSE53624 | NCK1 | 4690 | 83062 | -0.4085 | 0.0001 | |
| GSE63941 | NCK1 | 4690 | 211063_s_at | -0.4228 | 0.4868 | |
| GSE77861 | NCK1 | 4690 | 211063_s_at | -0.6309 | 0.1392 | |
| GSE97050 | NCK1 | 4690 | A_23_P255785 | 0.4225 | 0.3146 | |
| SRP007169 | NCK1 | 4690 | RNAseq | -0.3461 | 0.4067 | |
| SRP008496 | NCK1 | 4690 | RNAseq | -0.4505 | 0.0909 | |
| SRP064894 | NCK1 | 4690 | RNAseq | -0.6861 | 0.0046 | |
| SRP133303 | NCK1 | 4690 | RNAseq | -0.0344 | 0.8607 | |
| SRP159526 | NCK1 | 4690 | RNAseq | -0.2009 | 0.6270 | |
| SRP193095 | NCK1 | 4690 | RNAseq | -0.5235 | 0.0153 | |
| SRP219564 | NCK1 | 4690 | RNAseq | -0.6057 | 0.1390 | |
| TCGA | NCK1 | 4690 | RNAseq | 0.3172 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
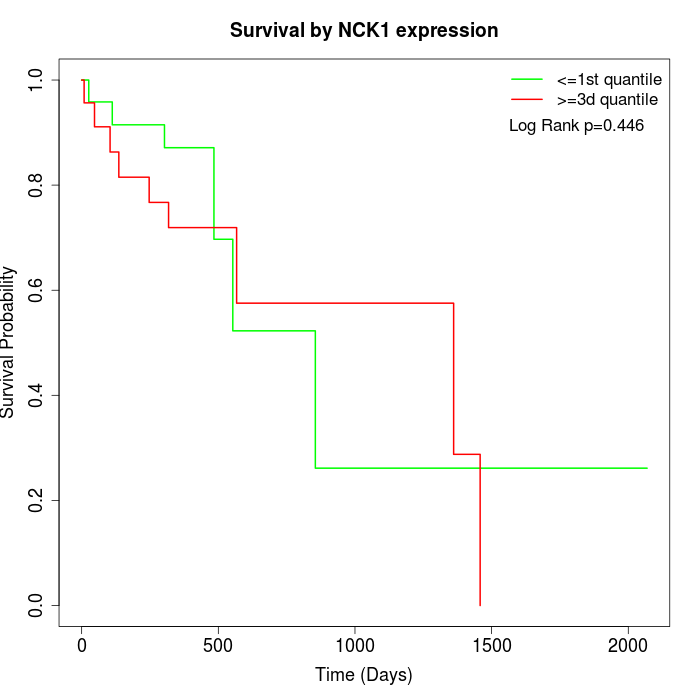
Survival by NCK1 expression:
Note: Click image to view full size file.
Copy number change of NCK1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NCK1 | 4690 | 16 | 0 | 14 | |
| GSE20123 | NCK1 | 4690 | 16 | 0 | 14 | |
| GSE43470 | NCK1 | 4690 | 22 | 0 | 21 | |
| GSE46452 | NCK1 | 4690 | 15 | 4 | 40 | |
| GSE47630 | NCK1 | 4690 | 18 | 3 | 19 | |
| GSE54993 | NCK1 | 4690 | 2 | 8 | 60 | |
| GSE54994 | NCK1 | 4690 | 32 | 1 | 20 | |
| GSE60625 | NCK1 | 4690 | 0 | 6 | 5 | |
| GSE74703 | NCK1 | 4690 | 19 | 0 | 17 | |
| GSE74704 | NCK1 | 4690 | 11 | 0 | 9 | |
| TCGA | NCK1 | 4690 | 60 | 6 | 30 |
Total number of gains: 211; Total number of losses: 28; Total Number of normals: 249.
Somatic mutations of NCK1:
Generating mutation plots.
Highly correlated genes for NCK1:
Showing top 20/423 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NCK1 | B4GALT5 | 0.818515 | 3 | 0 | 3 |
| NCK1 | ACSL5 | 0.722092 | 3 | 0 | 3 |
| NCK1 | NOTCH2 | 0.702493 | 3 | 0 | 3 |
| NCK1 | MOB4 | 0.702003 | 3 | 0 | 3 |
| NCK1 | CERS4 | 0.699434 | 3 | 0 | 3 |
| NCK1 | NDUFS1 | 0.693805 | 4 | 0 | 3 |
| NCK1 | SESN3 | 0.693043 | 3 | 0 | 3 |
| NCK1 | PLD2 | 0.689138 | 3 | 0 | 3 |
| NCK1 | DOCK10 | 0.686403 | 3 | 0 | 3 |
| NCK1 | HDHD2 | 0.685889 | 3 | 0 | 3 |
| NCK1 | CNOT6L | 0.680326 | 3 | 0 | 3 |
| NCK1 | STAM2 | 0.679668 | 4 | 0 | 4 |
| NCK1 | SLC30A5 | 0.674007 | 3 | 0 | 3 |
| NCK1 | PPP4R2 | 0.672504 | 3 | 0 | 3 |
| NCK1 | SH3BGRL3 | 0.670625 | 3 | 0 | 3 |
| NCK1 | WDR11 | 0.66966 | 3 | 0 | 3 |
| NCK1 | NAT1 | 0.66431 | 5 | 0 | 4 |
| NCK1 | FAM126B | 0.662963 | 4 | 0 | 3 |
| NCK1 | ELAVL1 | 0.662396 | 3 | 0 | 3 |
| NCK1 | TXNDC9 | 0.659479 | 4 | 0 | 3 |
For details and further investigation, click here