| Full name: nidogen 1 | Alias Symbol: entactin | ||
| Type: protein-coding gene | Cytoband: 1q42.3 | ||
| Entrez ID: 4811 | HGNC ID: HGNC:7821 | Ensembl Gene: ENSG00000116962 | OMIM ID: 131390 |
| Drug and gene relationship at DGIdb | |||
Expression of NID1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NID1 | 4811 | 202007_at | -0.5147 | 0.5438 | |
| GSE20347 | NID1 | 4811 | 202007_at | 1.3148 | 0.0027 | |
| GSE23400 | NID1 | 4811 | 202008_s_at | 0.1165 | 0.2979 | |
| GSE26886 | NID1 | 4811 | 202007_at | 2.7966 | 0.0000 | |
| GSE29001 | NID1 | 4811 | 202007_at | 1.0445 | 0.0401 | |
| GSE38129 | NID1 | 4811 | 202007_at | 0.8196 | 0.0992 | |
| GSE45670 | NID1 | 4811 | 202007_at | -0.8355 | 0.2748 | |
| GSE53622 | NID1 | 4811 | 5656 | 0.4272 | 0.0051 | |
| GSE53624 | NID1 | 4811 | 5656 | 0.8259 | 0.0000 | |
| GSE63941 | NID1 | 4811 | 202007_at | -4.8311 | 0.0172 | |
| GSE77861 | NID1 | 4811 | 1561082_at | 0.0465 | 0.5800 | |
| GSE97050 | NID1 | 4811 | A_33_P3281191 | 0.2021 | 0.7591 | |
| SRP007169 | NID1 | 4811 | RNAseq | 4.0763 | 0.0000 | |
| SRP008496 | NID1 | 4811 | RNAseq | 3.5530 | 0.0000 | |
| SRP064894 | NID1 | 4811 | RNAseq | 0.4944 | 0.1714 | |
| SRP133303 | NID1 | 4811 | RNAseq | 0.5124 | 0.1588 | |
| SRP159526 | NID1 | 4811 | RNAseq | 1.5627 | 0.0048 | |
| SRP193095 | NID1 | 4811 | RNAseq | 2.0430 | 0.0000 | |
| SRP219564 | NID1 | 4811 | RNAseq | -0.0123 | 0.9848 | |
| TCGA | NID1 | 4811 | RNAseq | -0.2183 | 0.0391 |
Upregulated datasets: 7; Downregulated datasets: 1.
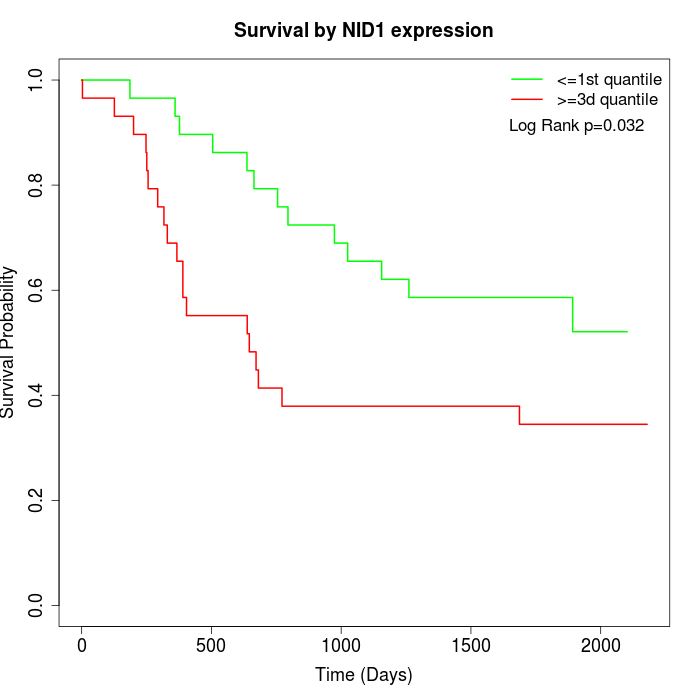
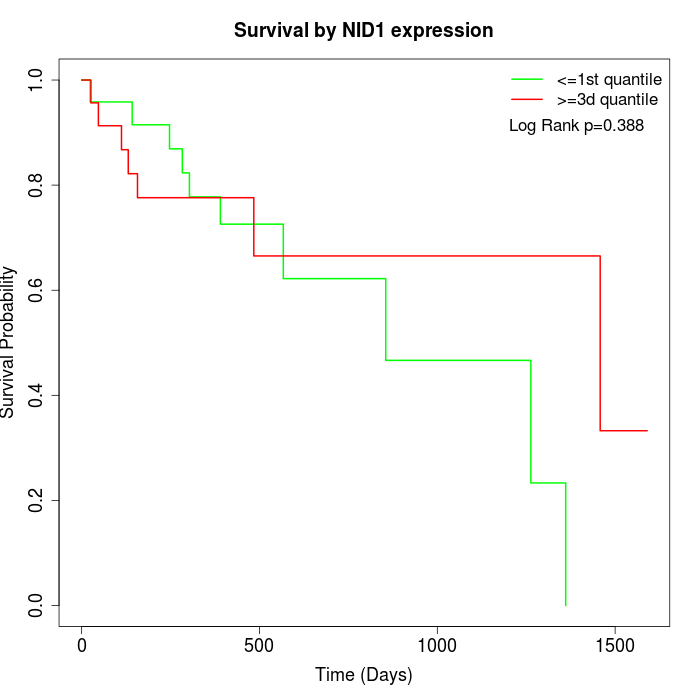
Survival by NID1 expression:
Note: Click image to view full size file.
Copy number change of NID1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NID1 | 4811 | 10 | 0 | 20 | |
| GSE20123 | NID1 | 4811 | 11 | 0 | 19 | |
| GSE43470 | NID1 | 4811 | 9 | 1 | 33 | |
| GSE46452 | NID1 | 4811 | 4 | 2 | 53 | |
| GSE47630 | NID1 | 4811 | 15 | 0 | 25 | |
| GSE54993 | NID1 | 4811 | 0 | 6 | 64 | |
| GSE54994 | NID1 | 4811 | 16 | 0 | 37 | |
| GSE60625 | NID1 | 4811 | 0 | 0 | 11 | |
| GSE74703 | NID1 | 4811 | 9 | 1 | 26 | |
| GSE74704 | NID1 | 4811 | 5 | 0 | 15 | |
| TCGA | NID1 | 4811 | 43 | 4 | 49 |
Total number of gains: 122; Total number of losses: 14; Total Number of normals: 352.
Somatic mutations of NID1:
Generating mutation plots.
Highly correlated genes for NID1:
Showing top 20/635 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NID1 | LAMA4 | 0.725847 | 10 | 0 | 10 |
| NID1 | COL12A1 | 0.721642 | 6 | 0 | 5 |
| NID1 | LAMC1 | 0.720859 | 10 | 0 | 9 |
| NID1 | FSTL1 | 0.71843 | 11 | 0 | 10 |
| NID1 | SDC2 | 0.704483 | 8 | 0 | 8 |
| NID1 | COL6A2 | 0.693596 | 9 | 0 | 9 |
| NID1 | FBN1 | 0.693401 | 9 | 0 | 8 |
| NID1 | RHOBTB1 | 0.690597 | 10 | 0 | 10 |
| NID1 | COL6A1 | 0.687313 | 10 | 0 | 10 |
| NID1 | CDC42EP3 | 0.686427 | 10 | 0 | 10 |
| NID1 | DOCK11 | 0.686065 | 5 | 0 | 5 |
| NID1 | IGFBP7 | 0.684389 | 10 | 0 | 9 |
| NID1 | COL6A3 | 0.683673 | 9 | 0 | 8 |
| NID1 | MMP2 | 0.677582 | 7 | 0 | 6 |
| NID1 | MICAL2 | 0.676859 | 10 | 0 | 9 |
| NID1 | COL4A2 | 0.676717 | 11 | 0 | 9 |
| NID1 | RHOQ | 0.673091 | 9 | 0 | 6 |
| NID1 | PMP22 | 0.671733 | 11 | 0 | 10 |
| NID1 | VCAN | 0.664068 | 9 | 0 | 8 |
| NID1 | MCAM | 0.663644 | 9 | 0 | 7 |
For details and further investigation, click here